Figure 5.
NKX2.5 binds a majority of ISL1 peaks and cooperates with ISL1 to regulate gene expression in CPs
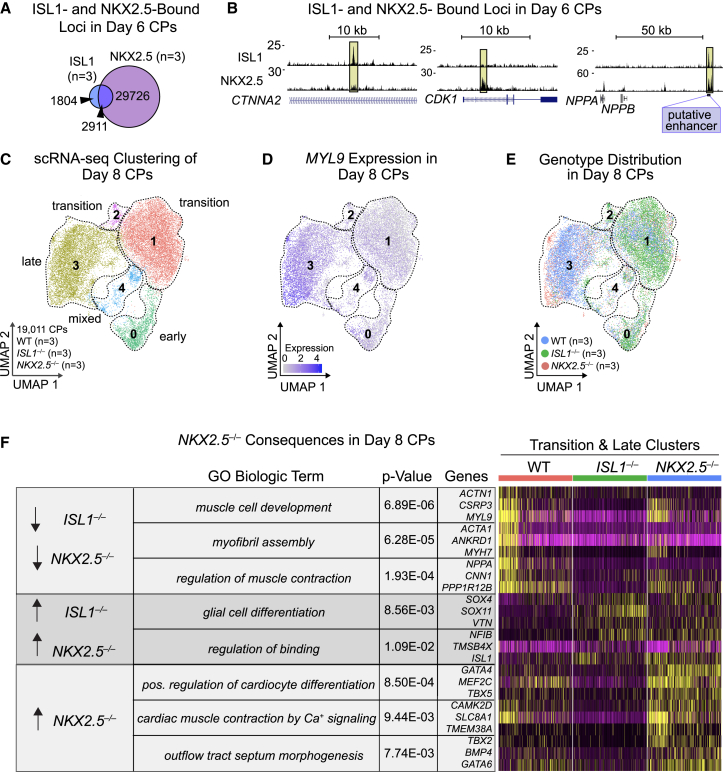
(A) Venn diagram of NKX2.5 and ISL1 ChIP-seq peaks in WT day 6 CPs.
(B) Example ChIP-seq tracks showing loci bound by ISL1, NKX2.5, or both in day 6 CPs. Putative NPPA/NPPB enhancer labeled as related to Figure S5I. Data shown from single representative replicate.
(C) Hierarchical UMAP scRNA-seq clustering of WT, ISL1−/−, and NKX2.5−/− day 8 CPs (n = 3 independent experiments).
(D) Expression of MYL9 in day 8 CPs, superimposed on the UMAP from (C).
(E) FeaturePlot display of the distribution of WT and ISL1−/− day 8 CPs across the clusters depicted in (C).
(F) Heatmap of significantly dysregulated genes with biological GO terms in day 8 ISL1−/− or NKX2.5−/− CPs compared with WT.