Figure 5.
Transcriptional changes are induced by suppression of neuronal activity in rat astrocytes
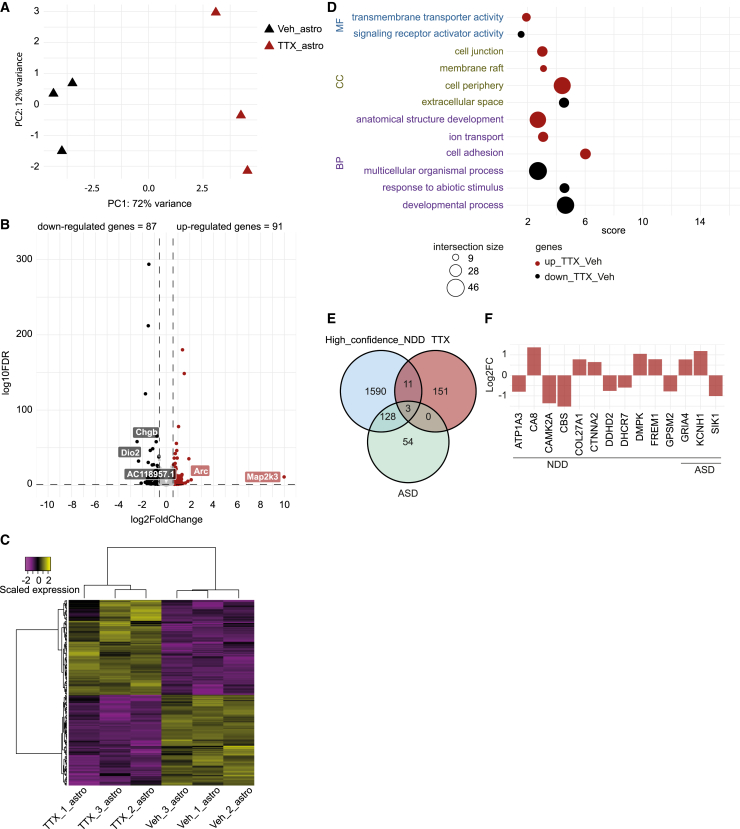
(A) PCA plot of RNA-sequencing data of six samples, three TTX- and three vehicle-exposed rat astrocytes derived from co-cultures with hiPSC-derived neurons (Ctrl1). Colors indicate the received treatment (red = TTX, and black = vehicle).
(B) Volcano plot depicting differential gene expression between TTX- and vehicle-exposed astrocytes. Colored dots indicate DEGs (Absolute Log2FC > 0.58 and adjusted p value <0.05). Up-regulated genes in TTX-exposed astrocytes are shown in red, down-regulated genes are depicted in black. The five genes with the greatest change in expression activity between the two conditions are indicated.
(C) Heatmap depicting scaled expression of DEGs in the six astrocyte samples.
(D) Scatterplot depicting the top three significant GO terms, per ontological category (molecular function: MF, cellular component: CC and biological process: BP), associated with up-regulated (red dots) and down-regulated genes (black dots) in astrocytes exposed to TTX treatment, as compared with vehicle-exposed astrocytes. The size of the dots indicates the number of intersected genes between the list of DEGs and the genes associated with the particular term. Score: negative logarithm10 of the adjusted p value resulting from the enrichment analysis.
(E) Venn diagram depicting the number of shared genes between the list of DEGs in TTX-exposed astrocytes (red circle, TTX) and genes previously related to NDDs (light-blue circle, High_confidence_NDD) (Leblond et al., 2021) and highly confident autism-related genes (light-green circle, ASD) (Fu et al., 2022).
(F) Bar plot depicting the Log2FC value of the 14 shared genes observed in (E). Fold change was calculated by normalizing gene expression level of TTX-treated condition to vehicle-treated condition.