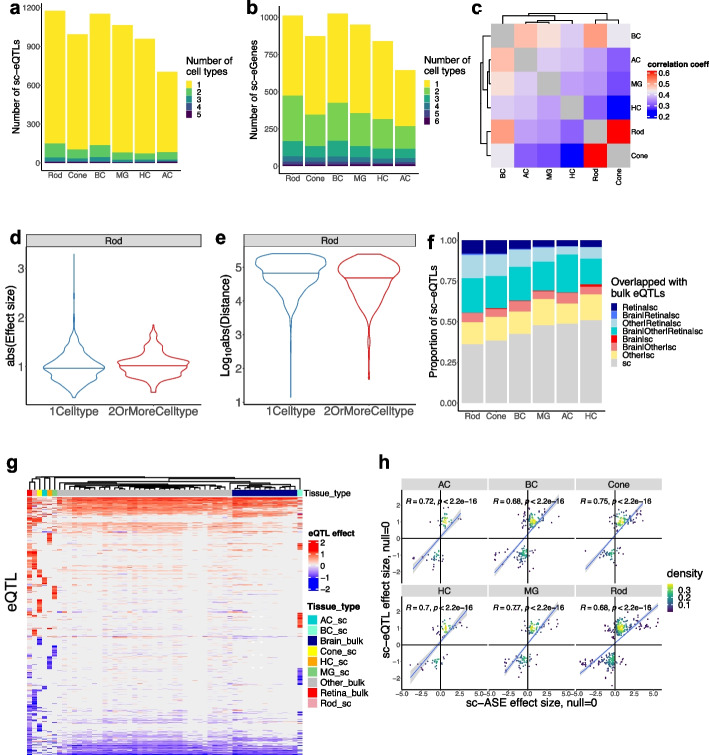
Fig. 2.
Identification of sc-eQTLs in retinal cell types. a Bar plot showing the number of independent index sc-eQTLs reaching gene-level FDR < 0.1 per cell type, colored by the number of cell types where a sc-eQTL is significant. b Bar plot showing the number of sc-eGenes reaching gene-level FDR < 0.1 per cell type, colored by the number of cell types where a sc-eGene is significant. c Heatmap showing the Pearson correlation of sc-eQTL effect size across retinal cell types. d Violin plot showing the distribution of absolute effect size of the sc-eQTLs located in non-promotor OCRs identified in Rod, colored by whether sc-eQTLs were identified in one (n = 1969) or more cell types (n = 539). To compare the effect size between the two types of sc-eQTLs, two-sided Wilcoxon rank sum test was performed, . e Violin plot showing the distribution of the distance between sc-eQTL and eGene TSS for the sc-eQTLs located in non-promotor OCRs identified in Rod, colored by whether sc-eQTLs were identified in one (n = 1969) or more cell types (n = 539). To compare the distance distribution between the two types of sc-eQTLs, two-sided Wilcoxon rank sum test was performed, . f Bar plot showing proportion of sc-eQTLs overlapping with bulk eQTLs, colored by the tissue types where the bulk eQTL is significant. sc: the identified sc-eQTLs. Other: other tissue. g Heatmap showing effect size of sc-eQTLs and the overlapped bulk eQTLs. Each row corresponds to an eQTL. Each column corresponds to a tissue or cell type. h Scatter plot showing the effect size of the overlapped sc-ASEs (X axis) and sc-eQTLs (Y axis) in the corresponding cell type. The Pearson correlation coefficient and p-value are indicated in the plot