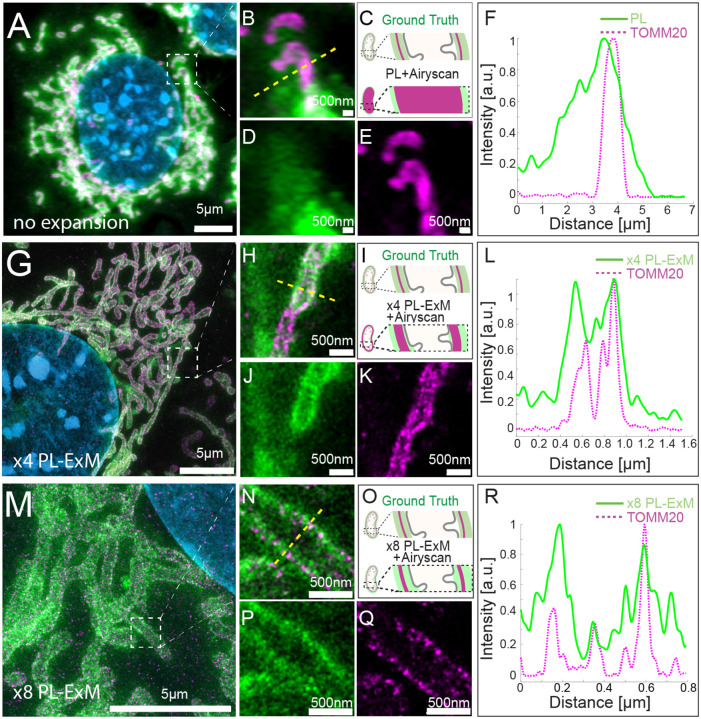
Figure2. PL-ExM offers super resolution for the visualization of the proximity-labeled interactome landscape.
All images were taken on MEF cells labeled with two colors in the same way. The TOMM20 was proximity-labeled to show its interactome (green) and simultaneously immunostained to locate the protein of interest (magenta). The nucleus was stained with DAPI (blue). All images were taken with Airyscan microscope. (A) Representative image of a non-expanded sample. (B) Magnified view of the boxed region in (A). (C) Schematics of the ground truth structure of proximity-labeled TOMM20 (green) and immunostained TOMM20 (magenta), and the expected image without expansion. (D) PL channel of (B). (E) Immunostained TOMM20 channel of (B). (F) A representative histogram showing the fluorescence intensity in a cross section of a mitochondrion from the image (B) of the non-expanded sample. The fluorescence intensity was denoised and normalized with respect to each channel. (G) Representative PL-ExM image of a 4-time expanded sample, named x4 PL-ExM. (H) Magnified view of the boxed region in (G). (I) Schematics of the same ground truth as in (C), and the expected PL-ExM image of the 4-time expanded sample. (J) PL-ExM channel of (H). (K) Immunostained TOMM20 channel of (H). (L) A representative histogram showing the fluorescence intensity in a cross section of mitochondrion from a x4 PL-ExM image. (M) Representative PL-ExM image of an 8-time expanded sample, named x8 PL-ExM. (N) Magnified view of the boxed region in (M). (O) Schematics of the same ground truth as in (C), and the expected PL-ExM image of the 8-time expanded sample. (P) PL-ExM channel of (N). (Q) Immunostained TOMM20 channel of (N). (R) A representative histogram showing the fluorescence intensity in a cross section of mitochondrion from an x8 PL-ExM image. In all histograms (F,L&R), the fluorescence intensity was denoised and normalized with respect to each channel. (A, G, M, N, P, Q) are maximum intensity projections of z stacks. (B, D, E, H, J, K) are single-slice images of 3D z stacks. Length expansion factors are 4.2 for samples (G, H, J, K), and 8.2 for (M, N, P, Q). All scale bars are in pre-expansion units.