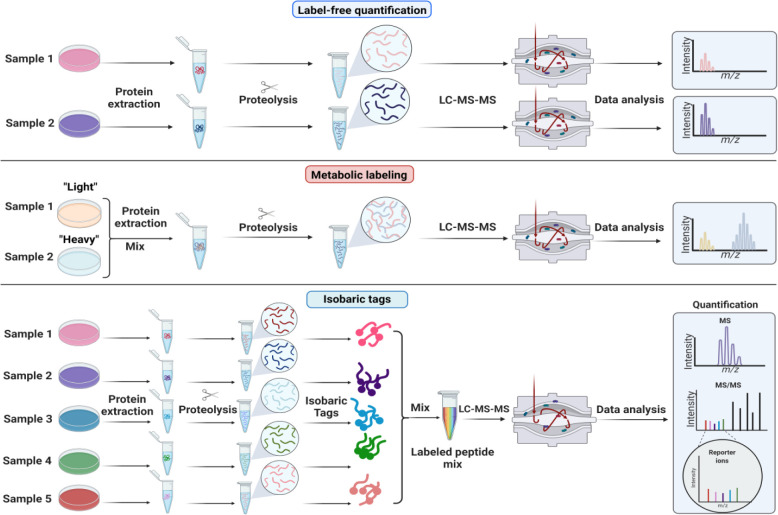
Figure 3: Quantitative strategies commonly used in proteomics.
A) Label-free quantitation. Proteins are extracted from samples, enzymatically hydrolyzed into peptides and analyzed by mass spectrometry. Chromatographic peak areas from peptides are compared across samples that are analyzed sequentially. B) Metabolic labelling. Stable isotope labeling with amino acids in cell culture (SILAC) is based on feeding cells stable isotope labeled amino acids (“light” or “heavy”). Samples grown with heavy or light amino acids are mixed before cell lysis. The relative intensities of the heavy and light peptide are used to compute protein changes between samples. C) Isobaric or chemical labelling. Proteins are isolated separately from samples, enzymatically hydrolyzed into peptides, and then chemically tagged with isobaric stable isotope labels. These isobaric tags produce unique reporter mass-to-charge (m/z) signals that are produced upon fragmentation with MS/MS. Peptide fragment ions are used to identify peptides, and the relative reporter ion signals are used for quantification.