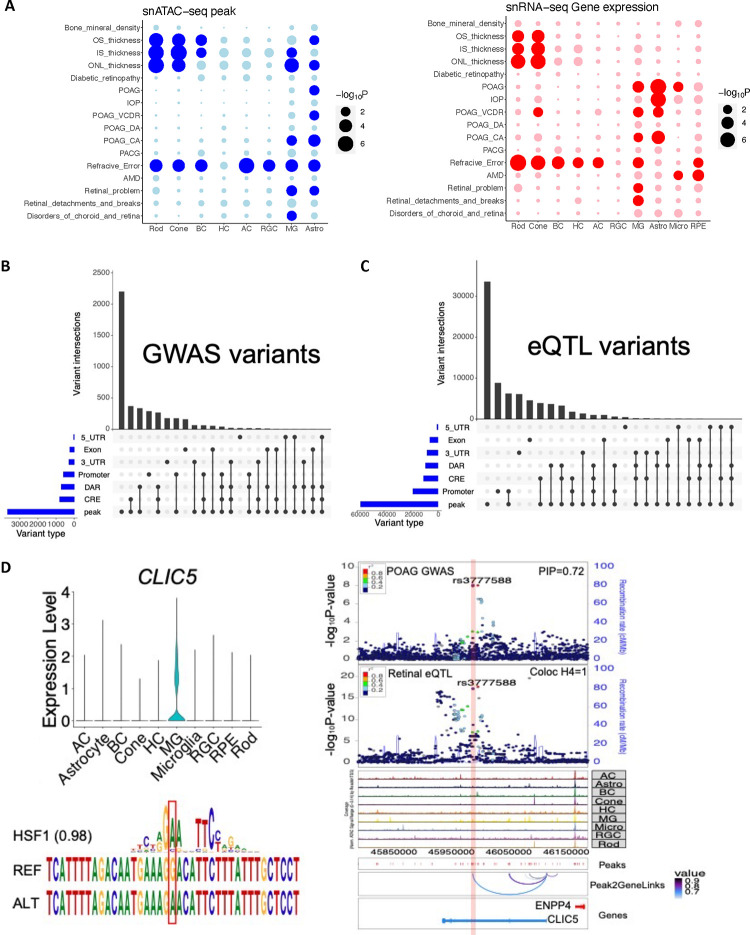
Figure 7. Leveraging multi-omics data to study GWAS and eQTL loci.
A. Cell class enrichment of GWAS loci based on chromatin accessibility with LDSC (left) and gene expression with MAGMA (right). Rows represent enriched GWAS traits, and columns represent retinal cell classes. The highlight dot indicates the enrichment q-value < 0.05. B. Categorization of fine-mapped GWAS variants located in various genomic regions. Categories include peak (i.e., open chromatin regions), linked cis-regulatory elements (CREs), differentially accessible regions (DARs), promoter, exon, 5_UTR and 3_UTR of gene annotation. C. Categorization of fine-mapped eQTL variants located in various genomic regions. D. Visualization of fine-mapped loci in CLIC5 region.