Figure 2. Missense rga mutant proteins showed varying degrees of reduced activities.
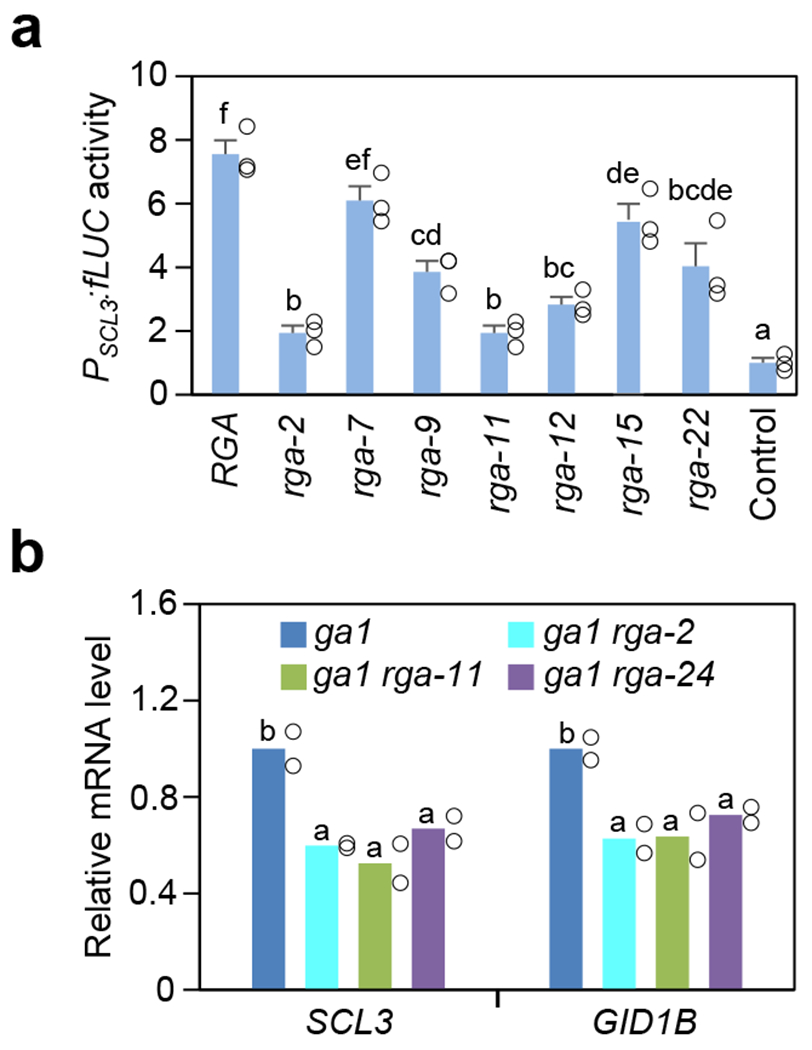
a, Dual luciferase assay in the N. benthamiana transient expression system showing rga mutant proteins were impaired in activating PSCL3:fLUC in comparison to RGA. Means ± SE of three biological replicates are shown. The reporter construct contained PSCL3:fLUC48. Effector constructs contained 35S:RGA or rga as labeled, and the empty vector was included as a negative control. RGA and rga proteins were expressed at similar levels in these assays (Supplementary Fig. 2a). b, RT-qPCR analysis showing rga-2 and rga-11 caused reduced expression of RGA-induced genes (SCL3 and GID1B) in planta, similar to the null rga-24 allele. The housekeeping gene, PP2A, was used to normalize different samples. Means of two biological replicates are shown. The level in ga1-3 was set to 1. In a and b, statistical analyses were performed with two-tailed (a) or one-tailed (b) Student’s t tests. Different letters above the bars represent significant differences, p < 0.05. Exact n and p values are listed in Source Data Fig. 2.
