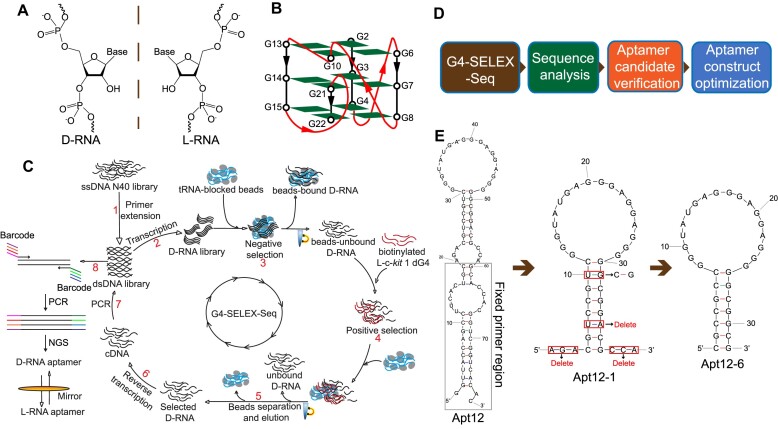
Figure 1.
Selection of L-RNA aptamer for c-kit 1 dG4 by G4-SELEX-Seq. (A) Chemical structures of D-RNA and L-RNA. (B) Schematic representation of the c-kit 1 parallel G-quadruplex structure. (C) Flowchart of G4-SELEX-Seq. D-RNA aptamers binding to the enantiomeric form of c-kit 1 dG4 target (L-c-kit 1 dG4) were first identified by in vitro selection. When the final D-RNA aptamer sequence was confirmed, L-RNA aptamer was then chemically synthesized to recognize D-c-kit 1 dG4 target. (D) Downstream processes after G4-SELEX-Seq to get an optimized aptamer. (E) Sequences and mFold predicted structures of full-length and truncated aptamers. The fixed primer region of full-length aptamer (Apt12) for G4-SELEX-Seq is boxed in gray. The N40 region (Apt12-1) was truncated by deleting the two flanking ends (5′-AGA and 3′-CCA) and the ‘U-A’ base pair in stem. To strengthen the stem region, the ‘U-G’ base pair originally in the stem region of Apt12-1 was mutated to ‘C-G’, and resulted in the final 32-nt aptamer sequence Apt12-6.