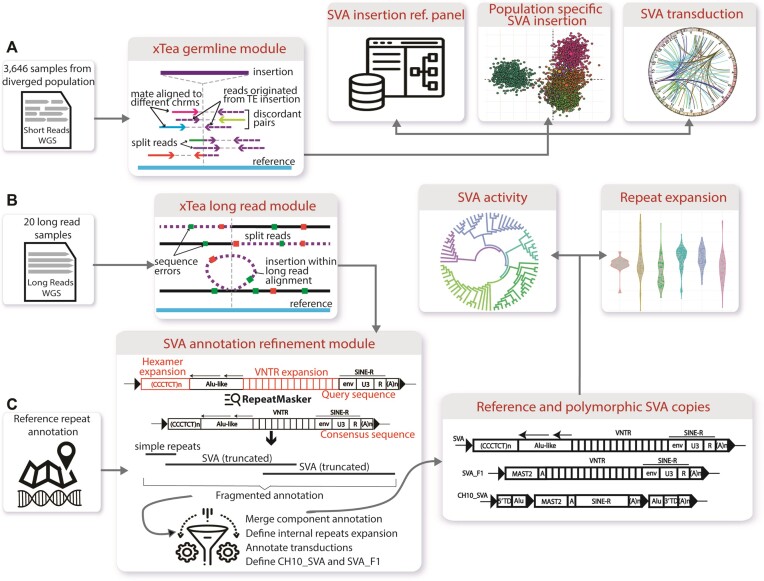
Figure 1.
SVA retrotransposon analysis workflow. (A) First, we run the xTea germline module on 3646 whole-genome samples. The integrated call set provides a comprehensive SVA reference map, defines population-specific SVA insertions, and identifies ‘hot’ source elements based on transductions. (B) In addition, we run the xTea long-read module on 20 Oxford Nanopore and PacBio long-read samples, and construct the full copies of both polymorphic and reference SVAs. (C) We developed a new module for SVA annotation. With refined annotation, we annotate the internal structure of the fully constructed SVAs, which allows us to characterize the distribution of hexamer and VNTR lengths and construct the SVA phylogeny tree to explore the SVA activity.