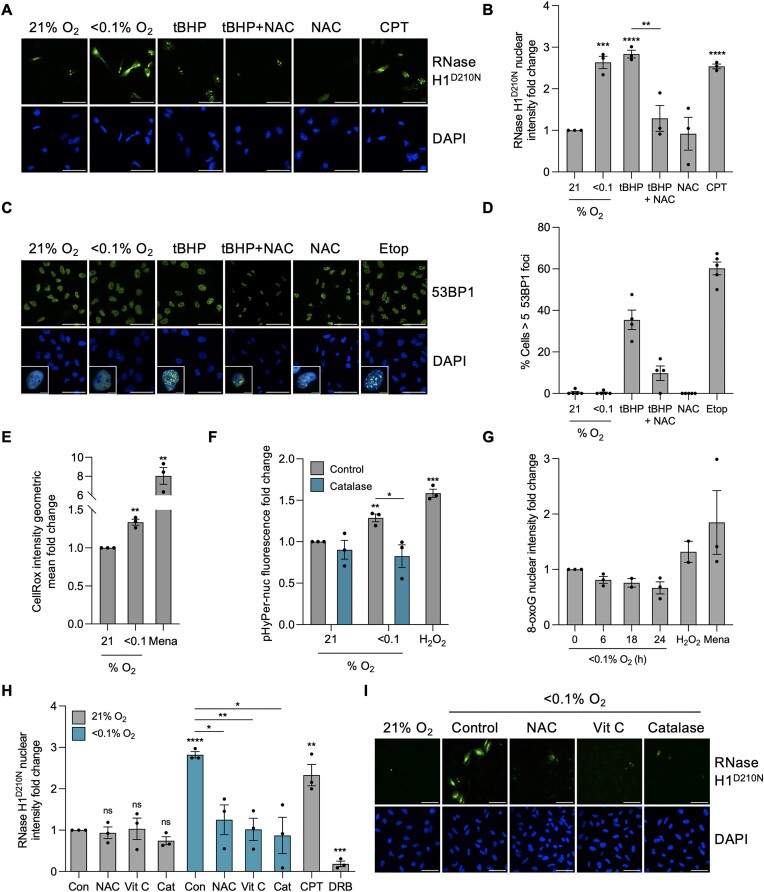
Figure 4.
R-loop accumulation in hypoxia (<0.1% O2) is ROS-dependent. (A) A549 cells were transfected with V5-tagged RNase H1D210N and exposed to 6 h of 21% O2, <0.1% O2, tBHP (20 μM), tBHP (20 μM) and NAC (20 mM), NAC (20 mM) or 1 h of CPT (10 μM). tBHP was used as a ROS-inducer, NAC was used as a ROS scavenger and CPT was used as a positive control for increasing R-loops. Cells were fixed and stained for V5 (green) and DAPI (blue). Scale bar represents 50 μM. (B) Quantification of V5-tagged RNase H1D210N nuclear intensity from part A. (C) A549 cells were exposed to 6 h of 21% O2, <0.1% O2, tBHP (20 μM), tBHP (20 μM) and NAC (20 mM), or NAC (20 mM). Etoposide (Etop, 25 μM, 6 h) was used as a positive control for DNA damage. Cells were fixed and stained for 53BP1. Scale bar represents 50 μM. n= 1. (D) Quantification of part (C). Each data point represents a field of view of cells where percentage of cells with >5 53BP1 foci were determined. At least 100 cells were imaged per condition. n= 1. (E) A549 cells were exposed to 21, <0.1 or 2% O2 (6 h) with CellRox (5 μM) added during the last 10 min of treatment. Cells were fixed and CellRox intensity was measured. Menadione (100 μM, 6 h) was used as a positive control to increase ROS. At least 10 000 cells were quantified per condition. (F) A549 cells were transfected with pHyPer-Nuc and exposed to 21, <0.1 or 2% O2 (6 h) with and without catalase (2000 U/mg). H2O2 (5 mM, 3 h) was used as a positive control. Cells were fixed and the nuclear intensity was determined. (G) A549 cells were exposed to 21 or <0.1% O2 for the times indicated. Cells were fixed and stained for 8-oxoguanine (8-oxoG), and the nuclear intensity was determined. H2O2 (5 mM, 3 h) and menadione (100 μM, 3 h) were used as positive controls. (H) A549 cells were transfected with V5-tagged RNase H1D210N, treated with or without NAC (20 mM), Vitamin C (2 mM) or catalase (2000 U/mg) then exposed to 21 or <0.1% O2 (6 h). Cells were fixed and stained for V5. (I) Representative images from part H. V5 (green) and DAPI (blue) are shown. Scale bar represents 50 μM. (A–I) Data from three independent experiments (n= 3), mean ± standard error of the mean (SEM) are displayed unless otherwise indicated. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, ns (non-significant) P > 0.05. Unless otherwise indicated statistical significance refers to comparison to the normoxic control. In parts (B), (E)–(H), each data point represents the average from one of three biological repeats, normalized to the untreated sample. A minimum of 100 cells was imaged per condition in all microscopy experiments. The two-tailed, unpaired Student's t-test was used in parts (B), (E), (F) and (H).