Abstract
Unscheduled R-loops are a major source of replication stress and DNA damage. R-loop-induced replication defects are sensed and suppressed by ATR kinase, whereas it is not known whether R-loop itself is actively involved in ATR activation and, if so, how this is achieved. Here, we report that the nuclear form of RNA-editing enzyme ADAR1 promotes ATR activation and resolves genome-wide R-loops, a process that requires its double-stranded RNA-binding domains. Mechanistically, ADAR1 interacts with TOPBP1 and facilitates its loading on perturbed replication forks by enhancing the association of TOPBP1 with RAD9 of the 9–1-1 complex. When replication is inhibited, DNA–RNA hybrid competes with TOPBP1 for ADAR1 binding to promote the translocation of ADAR1 from damaged fork to accumulate at R-loop region. There, ADAR1 recruits RNA helicases DHX9 and DDX21 to unwind R-loops, simultaneously allowing TOPBP1 to stimulate ATR more efficiently. Collectively, we propose that the tempo-spatially regulated assembly of ADAR1-nucleated protein complexes link R-loop clearance and ATR activation, while R-loops crosstalk with blocked replication forks by transposing ADAR1 to finetune ATR activity and safeguard the genome.
Graphical Abstract
Graphical Abstract.
Introduction
Replication stress in dividing cells is a major source of genome instability (1,2). Continuous activation of replication stress or failure to appropriately respond can induce functional exhaustion of hematopoietic stem and progenitor cells (HSPCs), causing senescence or apoptosis followed by bone marrow failure or malignancies (3,4). The protein kinase ataxia-telangiectasia mutated and Rad3-related (ATR), a master regulator of replication stress response, forms a stable complex with its obligatory partner ATR-interacting protein (ATRIP) in response to accumulation of single-stranded DNA (ssDNA) at stalled or collapsed replication forks (5,6). Once recruited to sites of stalled replication, ATR is activated by regulators including the RAD9-HUS1-RAD1 (9–1–1) complex and DNA topoisomerase 2-binding protein 1 (TOPBP1), a conserved multi-BRCA1 C-terminal (BRCT)-domain scaffolding protein (7,8). TOPBP1 interacts with phosphorylated RAD9 through its N-terminal BRCTs at ssDNA/double-stranded DNA (dsDNA) junctions, where it stimulates ATR’s kinase activity (8,9). ATR then phosphorylates replication protein A2 (RPA2) and its effector checkpoint kinase 1 (CHK1) to protect the genome and orchestrate cell cycle progression (10–12).
Transcription-replication collisions (TRCs) are a common source of genome instability, causing replication blockage and even collapse (13–17). When the actively transcribing RNA polymerase encounters a replication fork on the same DNA template, the nascent transcript may reanneal with the template DNA behind the RNA polymerase, leaving the non-coding ssDNA strand unpaired. This three-stranded structure, dubbed an R-loop, comprises a DNA–RNA hybrid, a stretch of displaced ssDNA, and a trailing ssRNA overhang (18,19). Tight regulation of R-loop generation, signaling, and resolution is critical for maintaining the physiological roles of R-loops (which include class-switch recombination, DNA repair, and transcription termination) and to avoid genome instability-associated diseases such as cancer and neurodegeneration (13,14,18,19). Many cellular factors are known to counteract R-loop accumulation: members of the RNase H family (RNH1 and 2) that degrade the DNA-hybridized RNA (20); helicases including DExH-box helicase 9 (DHX9) and DExD-box helicase 21 (DDX21) that unwind DNA–RNA hybrids (16,21–24); RNA-processing factors including serine/arginine-rich splicing factor 1 (SRSF1) (25,26); DNA repair factors (27–29); and chromatin modulators (30,31). An intimate interplay may constantly occur between R-loops and damaged replication forks – for example, the emergence of a significant fraction of stalled forks depends on DNA–RNA hybrids when ATR is depleted (32), and aberrant R-loop accumulation induced by splicing defect could activate the ATR-CHK1 pathway to promote replication fork recovery (26). Once replication is perturbed or at difficult-to-replicate regions, R-loops may form (13,15–17,32), whereas it remains unclear whether the resultant R-loop structure itself directly contributes to ATR activation at replication stress sites and how perturbed replication forks crosstalk with the concomitant R-loops.
Adenosine deaminase acting on RNA 1 (ADAR1) is a ubiquitously expressed RNA-editing enzyme that conducts adenosine-to-inosine (A-to-I) editing, a common RNA modification (33). ADAR1 exists in two different isoforms: a constitutively expressed nuclear protein (ADAR1p110) and an interferon-inducible cytosolic protein (ADAR1p150) (34). They share three repeated copies of a dsRNA-binding domain (RBD) and a single deaminase domain at the C-terminus (35). Mounting evidence demonstrates that ADAR1 is critical in maintaining immunological self-tolerance by editing cytosolic dsRNA (36,37) which would otherwise activate pattern recognition receptors including melanoma differentiation-associated gene 5 (MDA5) and protein kinase R (PKR). ADAR1 may also function independently of its editing activity by forming regulatory complexes with RNAs or proteins. For instance, ADAR1 coordinates with Dicer to promote microRNA processing and RNA-induced gene silencing (38), and it antagonizes Staufen1-mediated mRNA decay (39). Although loss of ADAR1 has been reported to decrease DNA damage response (40) or cause DNA damage and sensitize tumors to irradiation (41,42), far less clear is the underlying mechanism by which ADAR1 preserves genome stability. ADAR1 has been found to regulate R-loop formation at telomeres by its RNA editing activity (41), while it is still unknown that whether it could function beyond this specialized chromatin regions.
Here, we report that nuclear ADAR1 plays a critical, editing-independent role in sustaining genome integrity in mouse HSPCs and human cells by linking R-loop suppression to ATR activation. Once replication stress occurs, ADAR1 initially favors TOPBP1 loading and sets it in a poised state for ATR activation. The resultant R-loop from replication blockage then relocates ADAR1, allowing TOPBP1 to efficiently stimulate ATR. Consequently, ADAR1 accumulates and promotes R-loop removal by recruiting DNA–RNA hybrid unwinding factors. Our study reveals that ATR activity and R-loop homeostasis are coordinately regulated by ADAR1 and provides mechanistic insight into how R-loops contribute to ATR activation at replication stress sites.
Materials and methods
Antibodies and reagents
The sources of antibodies against the following proteins or post-translational modifications were as follows: RPA1 (2198S, for IF), γH2AX (9718S, for IF), phospho-CHK1 (Ser345) (2348S, for IB), ATM (2873, for IB) and ATRIP (2737T, for IF) from Cell Signaling Technology; and FLAG (F3165, IB, IF and IP) from Sigma-Aldrich; CHK2 (ab47433, for IB) and RPA2 (ab2175, for IF) from Abcam; TOPBP1 (A300-111A, for IB and IP), RAD9 (A300-890A, for IB), RPA2 pS33 (A300-246A, for IB and IF), Treslin (A303-472A, for IB) and biotin (A150-109A, for IB and IF) from Bethyl Laboratories; β-actin (AC004, for IB), DHX9 (A17955, for IB and PLA), DDX21 (A14820, for IB and PLA), PKR (A19545, for IB), FLAG (AE063, for IF and PLA), RAD1 (a1047, for IB) and HUS1 (a5407, for IB) from ABclonal; RNAPII pSer2 (NB100-1805, for PLA) from Novus Biologicals; ATR (19787–1-ap, for IB), CHK1 (25887–1-AP, for IB), RPA2 (10412–1-AP, for IB) and MDA5 (21775–1-AP, for IB) from Proteintech; ADAR1 (sc-73408, for IB and IP), PRPF19 (sc-514338, for IB), PCNA (sc-56, for PLA) and TOPBP1 (sc-271043, for IF) from Santa Cruz Biotechnology; IdU/BrdU (347580, for IF) from BD; CldU/BrdU (MCA2060GA, for IF) from AbD Serotec; GFP (YM3124, for IB and IP), Myc-tag (YM3002, for IB), His-tag (YM3004, for IB), and H2AX (YT2155, for IB) from ImmunoWay; and S9.6 (ENH001, for DRIP, IF, and PLA) from Kerafast. Glutathione Sepharose 4B (10260459) was purchased from GE Healthcare. Biotin-azide (B10184), Protein G Magnetic Beads (10004D) and Dynabead MyOne Streptavidin T1 (65601) were purchased from Invitrogen. EdU (900584), Anti-FLAG M2 affinity gel (A2220), 3 × FLAG peptide (F4799), His-Select Nickel Affinity Gel (P6611), CPT (C9911), HU (H8627), VE-821 (SML1415), IdU (I7125), CldU (C6891), blasticidin (15205), puromycin (P8833) and doxycycline (D9891) were purchased from Sigma-Aldrich. Cisplatin (S1166) was purchased from Selleck.
Plasmids
ADAR1p150 and ADAR1p110 cDNAs were chemically synthesized by You Bio and cloned into the pLenti-Puro-3 × FLAG-GFP or 3 × FLAG vector. Mutated and deleted variants of ADAR1 were generated by quick-change point mutation assays and PCR-based DNA recombination (G712R represents a variant with glycine replaced by arginine corresponding to G1007R in ADAR1p150, H615Q/E617A is a mutant with histidine to glutamine and glutamate to alanine substitution corresponding to H910Q/E912A in ADAR1p150, and EAXXA stands for a variant with the lysine residues of the KKXXK motif in all three RBDs replaced by glutamate or alanine). The GFP-tagged RNH1/Wt and RNH1/D210N were amplified from V5-tagged RNH1 constructs (kindly provided by Dr Liang Chen, Wuhan University, China) and integrated into the pLVX-Tight-Puro-nuclear localization signal (NLS)-GFP vector. The GST-tagged RBDs/Wt and RBDs/EAXXA were carried by the pGEX-6P-1 vector. The GFP-tagged wild type and BRCT domain deletions of TOPBP1, including Δ0–2, Δ3, Δ4–5, Δ6 and Δ7–8, were gifts from Pro. Miiko Sokka (Department of Biology, University of Eastern Finland, Finland). The 3 × FLAG-tagged TOPBP1 was amplified from GFP-TOPBP1 and integrated into the pLenti-Puro vector. His-tagged TOPBP1 BRCT variants were carried by a modified pET-28a-smt vector.
Cell culture
HeLa, U2OS, and HEK 293T cells were purchased from ATCC (Manassas, VA) and cultured under the manufacturer's instructions. LacO-LacI U2OS cells were kindly provided by Dr Roger Greenberg (University of Pennsylvania, Pennsylvania). Cells with doxycycline-inducible RNH1 expression were created in two steps. First, cells were infected with lentivirus carrying rtTA and subjected to blasticidin selection. Subsequently, the established rtTA cells were infected with a virus carrying a pLenti-Tight-Puro vector that encodes RNH1 variants, followed by puromycin selection. Cells with integrated rtTA were cultured in Tet System Approved FBS and medium from Clontech. All of the cells were authenticated by examination of morphology and growth characteristics and confirmed to be mycoplasma-free.
Mice
Adar1 fl/fl mice on a C57BL/6 background bred inhouse (43) were crossed to B6.Cg-Tg(CAG-cre/Esr1)5Amc/J (ER-Cre) purchased from Jackson Laboratory. c-Kit+ hematopoietic stem and progenitor cells (HSPCs) were isolated from 16-week old mice harboring floxed Adar1 alleles (Adar1fl/fl) and ER-Cre and were cultured in the presence of 4-OHT (10 μM) for 2 days to excise the floxed Adar1 alleles. All procedures involving animals were approved by the Institutional Animal Care and Use Committees of the State Key Laboratory of Experimental Hematology and followed the NIH Guide for the Care and Use of Laboratory Animals (8th ed. The National Academies Press, 2011).
Colony-forming unit (CFU) assay
HSPCs were cultured in the presence or absence of 4-OHT for 48 h, then plated on 12-well plates in methylcellulose medium (MethoCult GF M3434; STEMCELL Technologies) at a density of 1000 cells/ml. The medium was supplemented with 1% penicillin/streptomycin (Gibco/Thermo Fisher Scientific). The cells were incubated at 37°C with 5% CO2 for 10–14 days to generate colonies.
Protein expression and purification
The His-tagged BRCTs expressed in the BL21 (DE3) strain of E. coli were induced with 0.15 mM IPTG at 16°C overnight. Cells were harvested by centrifugation and sonicated in buffer A (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM imidazole, 1 mM EDTA, 1 mM β-mercaptoethanol). Cell debris was removed by centrifugation at 4000 g for 40 min at 4°C. The supernatant of His-tagged proteins was loaded onto a Ni Sepharose Excel Column (Cytiva), followed by washing the beads with buffer A. The fusion protein was eluted with buffer B (25 mM Tris–HCl pH 8.0, 500 mM NaCl, 200 mM imidazole, 1 mM EDTA, and 1 mM β-mercaptoethanol). The protein was then loaded onto a HiTrap SP HP column (Cytiva) pre-equilibrated with buffer C (20 mM MES pH 6.0, 100 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol) and eluted with a linear gradient of 0.1–1.0 M NaCl. The eluted protein was concentrated by ultrafiltration and further purified using a HiLoad Superdex 200 16/60 size-exclusion column (GE Healthcare) in a buffer containing 20 mM MES pH 6.0, 300 mM NaCl and 1 mM β-mercaptoethanol. High-purity fractions were pooled, concentrated, and stored in PBS buffer. Similarly, GST-tagged RBDs were purified from BL21 E. coli with Glutathione Sepharose 4B (GE Healthcare), and the eluted proteins were then concentrated by ultrafiltration and further purified by HiTrap Heparin HP (Cytiva) and HiLoad Superdex 200 16/60 columns.
RNA interference
All siRNA transfections were performed using Lipofectamine RNAi MAX (Invitrogen) following the manufacturer's recommendations. The final concentration of the siRNA molecules was 5 nM, and cells were harvested 72 h later. Control siRNA (ON-TARGETplus Non-Targeting Pool, D-001810–10) was purchased from Dharmacon, and the other siRNAs were chemically synthesized by Sigma-Aldrich (Shanghai, China). The siRNA sequences are listed in Supplementary Table S1.
Lentiviral production
The lentivirus vectors encoding ADAR1, RNH1, and their corresponding variants were individually co-transfected with three assistant vectors – pMDLg/pRRE, pRSV-REV, and pVSV-G into HEK 293T cells. Viral supernatants were collected 48 h later, clarified by filtration, and concentrated by ultracentrifugation. The lentivirus was then used to transduce cells, followed by antibiotic selection to generate stable lines.
Co-IP
Cell lysates were prepared by incubating cells in NETN buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.2% Nonidet P-40, 2 mM EDTA) in the presence of Protease Inhibitor Cocktails (Roche) for 20 min at 4°C. This was followed by centrifugation at 14000 g for 15 min at 4 °C. For IP, ∼500 μg of protein was incubated with control or specific antibodies (2 μg) for 12 h at 4°C with constant rotation; 50 μl of 50% protein G magnetic beads (Invitrogen) were then added, and the incubation was continued for an additional 2 h. Beads were then washed five times using the NETN buffer. Between washes, the beads were collected by a magnetic stand (Invitrogen) at 4°C. The precipitated proteins were eluted from the beads by re-suspending in 2 × SDS-PAGE loading buffer and boiling for 5 min. The boiled immune complexes were subjected to SDS-PAGE followed by immunoblotting with appropriate antibodies. When examining the role of phosphorylation in ADAR1-TOPBP1 binding, HeLa cell extracts were pre-treated with λ-phosphatase (8 U/μl) for 0.5 h at 37°C before IP.
GST pull-down assays
GST-fusion RBDs were incubated with His-tagged BRCTs at 4°C for 2 h followed by the addition of Glutathione Sepharose 4B beads. After incubating with beads for 1 h at 4°C, the beads were washed 4 times, then boiled in SDS loading buffer and subjected to SDS-PAGE followed by Coomassie brilliant blue staining or immunoblotting. For the competitive binding experiment, the preformed RBDs-BRCT 0–2 complex was challenged with different amounts of the hybrids at 4°C overnight in binding buffer (20 mM HEPES pH 7.5, 5 mM magnesium acetate, 1 mM dithiothreitol, 0.5% NP-40, 10% [v/v] glycerol, 150 mM NaCl, 1 mg/ml BSA). After washing, the resultant products were examined by immunoblotting.
Immunopurification and silver staining
Lysates from HeLa cells stably expressing control vector or FLAG-ADAR1 were collected by incubating the cells in lysis buffer containing Protease Inhibitor Cocktail (Roche). Anti-FLAG immunoaffinity columns were prepared using anti-FLAG M2 affinity gel following the manufacturer's suggestions. Cell lysates were obtained from ∼5 × 108 cells and applied to an equilibrated FLAG column of 1 ml bed volume to allow for adsorption of the protein complex to the column resin. After binding, the column was washed with cold PBS + 0.2% Nonidet P-40. FLAG peptide was applied to the column to elute the FLAG protein complex flowing a protocol described by the vendor. The eluents were collected and visualized on SDS-PAGE followed by silver staining with a silver stain kit (Pierce).
Mass spectrometry (MS) analysis and data processing
Proteins were separated by SDS-PAGE and visualized by silver staining. The corresponding bands were then excised and subjected to in-gel digestion. The resulting peptides were desalted, redissolved in HPLC buffer A (0.1% formic acid in water), and injected into a Nano-LC system (EASY-nLC 1200, Thermo Fisher Scientific). Peptides were separated using a reversed-phase analytical column and electrosprayed directly into an Orbitrap Q-Exactive Plus mass spectrometer. The MS analysis was carried out in a data-dependent mode with an automatic switch between a full MS scan and an MS/MS scan in the orbitrap. For the full MS survey scan, the automatic gain control (AGC) target was 1e6 and scan range was from 350 to 1750 with a resolution of 70 000. The ten most intense peaks with charge state 2 and above were selected for fragmentation by higher-energy collision dissociation (HCD) with normalized collision energy of 27%. The MS2 spectra were acquired with 17 500 resolution. The exclusion duration for the data-dependent scan was 10 s, and the exclusion window was set at 1.6 Da. The resulting MS/MS data were searched using Proteome Discoverer software (v1.4) with an overall false discovery rate (FDR) for peptides of less than 1%. Proteins demonstrating the score < 2 and single-peptide identifications were removed from identification list. Peptide sequences were searched against the UniProt human database using trypsin specificity and allowing a maximum of two missed cleavages. Carbamidomethylation on Cysteine was specified as fixed modification. Oxidation of methionine and acetylation on peptide N-terminal were set as variable modifications. Mass tolerances for precursor ions were set at ±10 ppm for precursor ions and ±0.02 Da for MS/MS.
Immunofluorescence
Cells were seeded on glass coverslips (BD Biosciences), fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100 in PBS. Samples were blocked in 5% donkey serum in the presence of 0.1% Triton X-100 and stained with the appropriate primary and secondary antibodies coupled to Alexa Fluor 488, 594 or 647 (Invitrogen). The murine c-Kit+ HSPCs were centrifuged onto glass coverslips at 800 g for 5 min, followed by fixation and immunostaining as above. Confocal images of regular scan area or broad area were captured on a Zeiss LSM 900 microscope with a 63× oil objective. To avoid bleed-through effects in double-staining experiments, each dye was scanned independently in a multi-tracking mode. When inspecting nuclear-wide dispersed RPA2, TOPBP1, or RPA2 pS33 foci, cells were pre-treated with 0.5% Triton X-100 for 5 min on ice to extract non-chromatin fractions and fixed with 3% paraformaldehyde and 2% sucrose for 15 min at room temperature. Cells were then permeabilized with 0.5% Triton X-100 for 5 min on ice and incubated in blocking buffer (0.1% Triton X-100 and 5% donkey serum in PBS) for 1 h at room temperature. For S-phase discrimination, cells were pulsed with 10 μM EdU at 37°C for 1 h before fixation. Incorporated EdU was click-labeled using keyFluor 647-azide (Keygen Technologies) according to the manufacturer's instructions. For immunofluorescence with S9.6 antibody staining, cells were harvested, washed, and resuspended with PBS. Pre-warmed 75 mM KCl solution was added dropwise while the cells were gently shaken, followed by incubation at 37°C for 12 min. Afterwards, freshly made, ice-cold methanol:acetic acid (3:1) was added dropwise. Cells were then fixed on ice in methanol:acetic acid for 20 min followed by spotting onto slides. Slides were air-dried and immediately treated with blocking buffer (5% BSA and 0.5% Triton X-100 in PBS) for 1 h at room temperature. A standard immunostaining procedure was then performed with the primary antibody (S9.6) and the appropriate secondary antibodies conjugated with Alexa Fluro 594 (Invitrogen). The foci number or intensity of R-loops, γH2AX, RPA2, TOPBP1 and RPA2 pS33 was quantified using ImageJ software (NIH).
Proximity ligation assay (PLA)
Cells on glass coverslips were washed once with cold PBS and then treated with cold CSK extraction buffer (0.2% Triton X-100, 20 mM HEPES–KOH pH 7.9, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 1 mM EGTA) on ice for 3 min. Next, cells were fixed on ice with 4% paraformaldehyde for 5 min, then blocked with 10% FBS in PBS for 1 h at 37°C in a humidity chamber. Cells were washed with PBS between each step. Afterward, cells were incubated with the indicated primary antibodies at 4°C overnight. PLA was performed according to the manufacturer's instructions with Duolink In Situ Detection Reagents (DUO92101, Sigma-Aldrich). Finally, coverslips were mounted on slides with Fluoroshield (Sigma-Aldrich) containing DAPI. Images were captured with a Zeiss LSM 900 microscope using a 63× oil objective and quantified using ImageJ.
In situ analysis of protein interactions at DNA replication forks (SIRF)
Cells were grown on glass coverslips and labeled with 125 μM EdU for 8 min before treatment with 1 mM HU for 4 h. Cells were then washed with cold PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.25% TritonX-100 for 15 min. After permeabilization, slides were washed twice with cold PBS and Click-iT assay was performed in reaction buffer (10 μM biotin-azide, 2 mM CuSO4, and 100 mM sodium ascorbate) for 1 h at 37 °C. The cells were then washed twice with PBS and incubated with blocking buffer (10% BSA and 0.1% Triton X-100 in PBS) for 1 h at 37 °C, followed by PLA assays with primary antibodies against biotin and proteins of interest.
Reconstitution of DNA–RNA hybrid and R-loop
RNA oligos were 5′-labeled with Cy3. RNA and DNA oligos were annealed to generate a hybrid or R-loop structure by heating to 95°C for 5 min and slow cooling with a touch-down program over night in buffer H (90 mM Tris–HCl pH 7.5, 10 mM MgCl2, 50 mM NaCl). Annealed substrates were separated on a 6% native PAGE in 0.5 × TBE buffer (45 mM Tris-borate pH 8.0, 1 mM EDTA) at 4°C. The gel band corresponding to the annealed substrate was excised, purified, and finally eluted. The reconstituted substrates were concentrated and used in gel electrophoresis and competition assays. The DNA and RNA oligo sequences are listed in Supplementary Table S2.
Electrophoretic mobility shift assay (EMSA)
DNA–RNA hybrid (90 nt) or R-loop containing Cy3-labeled RNA were incubated with GST-tagged RBDs at 4°C overnight in binding buffer (20 mM HEPES pH 7.5, 5 mM magnesium acetate, 1 mM dithiothreitol, 0.5% NP-40, 10% glycerol, 150 mM NaCl, 1 mg/ml BSA). The resulting protein-substrate complexes were resolved on 6% polyacrylamide gels using 0.5× TBE buffer. Signals were detected with a fluorescence image analysis system (Tanon 5200 Multi), and the band intensities were quantified using ImageJ.
Nuclear extracts-based ATR activation assay
Nuclear extracts from HeLa cells were supplemented with reaction buffer (10 mM HEPES, pH 7.6, 50 mM KCl, 0.1 mM MgCl2, 1 mM PMSF, 0.5 mM DTT, 1 mM ATP, 10 mg/ml creatine kinase, 5 mM phosphocreatine). Blocked replication fork analogue was incubated with the nuclear extracts for 15 min at 37°C, after which DNA–RNA hybrid (25 nt) was added and the reaction was stopped 1 h later. The reaction mixtures were then boiled and analyzed by immunoblotting.
DNA–RNA Hybrid IP (DRIP) and DRIP-seq
Cells were lysed in 85 mM KCl, 5 mM PIPES pH 8.0, and 0.5% NP-40 for 10 min on ice. Nuclei pellets were collected and resuspended in buffer containing 10 mM Tris–HCl pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.2% sodium deoxycholate, 0.1% SDS, 0.05% sodium lauroyl sarcosinate, and 0.5% Triton X-100 to extract chromatins. The chromatins were then sonicated to ∼200 bp and diluted five-fold in dilution buffer (10 mM Tris–HCl pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100). S9.6 antibody was added to the fragmented chromatins for immunoprecipitation at 4°C overnight, and the incubation was continued with protein G dynabeads (Invitrogen) for 2 h. RNase A (PureLink, Invitrogen) was added during immunoprecipitation at a concentration of 0.1 ng RNase A per μg genomic DNA. The dynabead-conjugated chromatins were washed 4 times with dilution buffer and twice with washing buffer (10 mM Tris–HCl pH 7.5, 200 mM NaCl, 2.5 mM MgCl2), followed by immunoblotting. For DRIP-seq, the DNA–RNA hybrids were fragmented to ∼500 bp on average by enzymatic digestion followed by sonication and purified for library construction with a VAHTS Universal DNA Library Prep Kit for Illumina V3 according to the manufacturer's instructions (ND607, Vazyme). Finally, the prepared library was used for paired-end sequencing (PE150) on a NovaSeq platform. The clean reads of DRIP-seq data were aligned to Human genome reference (hg19 assembly) with Bowtie2 (version 2.4.5), and peak-calling was done using MACS2 with a q-value of 0.005. R-loop-gained peaks in ADAR1 depleted cells were determined as those with a fold-change >1.5 and P < 0.0001. R-loop levels on intron 1 of ACTB and control genomic region were analyzed by SYBR Green-based (TransGen Biotech) quantitative PCR (qPCR). The qPCR primers with single melting curves are provided in Supplementary Table S3.
Dot blotting
DNA samples were extracted via QIAamp DNA mini kit (QIAGEN) and digested with RNase H (NEB) at 37°C for 1 h. The equal amounts of DNA was then spotted on a positively charged nylon transfer membrane (Roche) and dried at room temperature. Next, the membrane was UV crosslinked (120 mJ/cm2) and blocked with 5% milk in PBST buffer (0.1% Triton X-100 in PBS) at room temperature for 1 h. The membrane was incubated with S9.6 antibody or dsDNA antibody in blocking buffer, followed by the addition of secondary antibodies coupled with horseradish peroxidase (HRP). Blot signals were detected by chemiluminescence. To examine the telomeric DNA–RNA hybrids, the DRIP products and 5% of input genomic DNA were spotted onto a Hybond N+ nylon filter. The membrane with spotted DNA was UV cross-linked and washed in 5× saline sodium citrate (SSC, B548110-0200) for 1 min after denaturation with 0.5 M NaOH for 2 min. The membrane was then prehybridized with Ultrasensitive Hybridization Buffer (Invitrogen, AM8670) overnight at 42°C. Biotin-labeled telomeric probes were incubated with the membrane for 10–12 h in hybridization buffer at 42°C. After three washes with 2× SSC/0.1% SDS solution for 15 min at 42°C, the membrane was blocked with 5% skim milk in TBST for 1 h at room temperature. The membrane was incubated with biotin antibody in blocking buffer overnight at 4°C, followed by incubation with HRP-conjugated secondary antibody and chemiluminescence detection. The sequences of two types of telomeric probes are shown in Supplementary Table S4.
Comet assay
The CometAssay kit (Trevigen, Gaithersburg, MD, USA) was used to monitor damaged DNA according to the manufacturer's instructions. Briefly, cells were re-suspended in ice-cold PBS at a concentration of 1 × 105 cells/ml. Cells (5 μl) were mixed with 50 μl of warm low-melting agarose and evenly spread onto the comet slides. Slides were then incubated in a pre-chilled lysis solution for 60 min at 4°C. Next, the slides were incubated with neutral unwinding solution for 60 min at room temperature. The slides were transferred to an electrophoresis tank containing a pre-chilled neutral electrophoresis solution, and the system was run at 1 volt/cm, 300 mA for 30 min at 4°C. The slides were immersed twice in deionized water for 5 min and washed in 70% ethanol for 5 min. Cells were then stained with 100 μl propidium iodide for 20 min in the dark and analyzed using an Olympus IX71 inverted fluorescence microscope. Olive tail moment was calculated with the formula: (tail mean-head mean) × % of DNA in the tail.
Cell survival assay
Cells were plated into 96-well plates at a density of 2000 cells/well. After 24 h, cells were treated with various doses of genotoxic agents for 72 h. CellTiter AQueous One Solution (Promega) was then added to each well according to the manufacturer's instructions. Cell survival was determined after 1 h incubation by measuring the absorbance at 490 nm using a Bio-Rad plate-reader (Model 550; Bio-Rad).
Statistics
Data from biological triplicate experiments are presented as means ± SDs. Two-tailed unpaired Student's t-test was used for comparing two groups of data. Analysis of variance (ANOVA) with the Bonferroni correction was used to compare multiple groups of data. For values not normally distributed, Mann–Whitney U test or Kruskal–Wallis test was used. P < 0.01 was considered statistically significant. All statistical analyses involved using Statistical Product and Service Solutions (SPSS, version 19.0.0). Before statistical analysis, variation within each group of data and the assumptions of the tests were checked.
Results
Nuclear ADAR1 is required for genome integrity
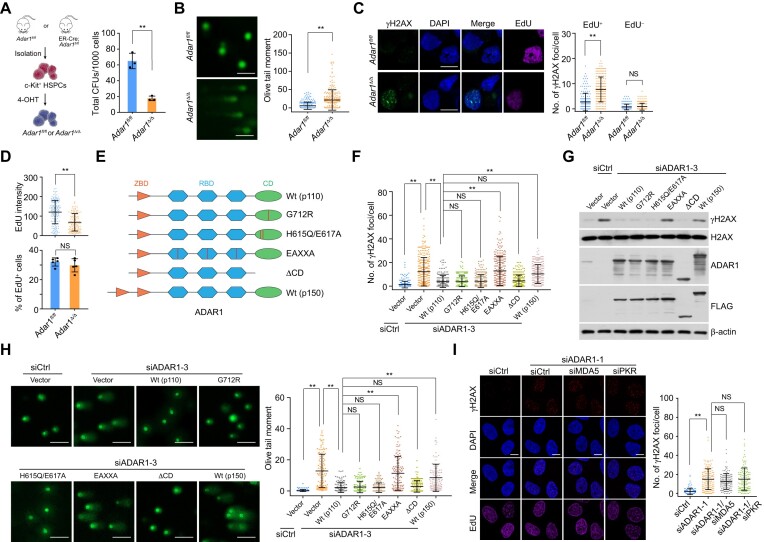
Erroneous response to replication stress is believed to be a potent driver of functional loss in HSPCs (4), and we previously found that apoptosis increases in ADAR1-deficient HSPCs (43). To determine whether this phenomenon is due to genome instability, we generated Adar1-knockout HSPCs from ER-Cre;Adar1fl/fl mice (Figure 1A and S1A) and confirmed that ADAR1 deletion significantly inhibited HSPC colony formation (Figure 1A). Immunoblotting showed that γH2AX, a chromatin marker that reflects endogenous genome instability (44), was upregulated in Adar1-knockout HSPCs (Supplementary Figure S1B), which also accumulated damaged DNA as evidenced by the alkaline comet assay (Figure 1B). Next, to evaluate the role of ADAR1 in actively replicating HSPCs, we examined the formation of γH2AX foci in EdU-labeled cells, finding a marked increase in the number of γH2AX foci in Adar1-knockout EdU-positive HSPCs (Figure 1C). EdU fluorescence intensity (though not the overall fraction of EdU-positive cells) was also diminished in Adar1-knockout cells, suggesting DNA replication defects (Figure 1D). These results indicate that ADAR1 plays a role of importance in preserving genome stability in actively replicating HSPCs.
Figure 1.
Nuclear ADAR1 is required for genome integrity. (A) Colony formation of control (Adar1fl/fl) and Adar1-knockout (Adar1Δ/Δ) HSPCs. A schematic illustrating the generation of Adar1-knockout HSPCs (c-Kit+ bone marrow cells) is also shown. (B) Analysis of damaged DNA accumulation by alkaline comet assay in control and Adar1-knockout HSPCs. Olive tail moment in single cell gel electrophoresis was calculated according to the tail length and the fraction of total DNA in the tail (n > 220 from 5 mice). Scale bars, 100 μm. (C) Immunostaining followed by confocal microscopy analysis of γH2AX foci formation in EdU-labeled control and Adar1-knockout HSPCs. Cells were labeled with EdU for 1 h before collection. The number of γH2AX foci per cell is shown (n > 160 from 5 mice). Scale bars, 10 μm. (D) Quantification of EdU intensity and the percentage of EdU-positive cells in control and Adar1-knockout HSPCs. The same data in (C) were re-analyzed. (E) A schematic of ADAR1 domains and its mutation or deletion mutants. ZBD, Z DNA binding domain; RBD, double-stranded RNA binding domain; CD, catalytic domain. (F) Immunostaining followed by confocal microscopy analysis of γH2AX foci formation in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated stably integrated ADAR1 variants (n > 200). Scale bars, 10 μm. (G) Immunoblotting analysis of γH2AX levels and expression levels of the indicated proteins in HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants. (H) Analysis of damaged DNA accumulation by alkaline comet assay in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated stably integrated ADAR1 variants (n > 100). Scale bars, 100 μm. (I) Immunostaining followed by confocal microscopy analysis of γH2AX foci formation in HeLa cells expressing the indicated siRNAs (n > 110). Scale bars, 10 μm. Data are mean ± SDs for (A–D), (F) and (H, I) from biological triplicate experiments. **P < 0.01; NS, not significant; two-tailed unpaired Student's t-test for (A) and (D, lower), Mann–Whitney test for (B) and (D, upper), and Kruskal–Wallis test for (C), (F) and (H, I).
We next knocked down ADAR1 using distinct siRNAs against all ADAR1 isoforms in HeLa cells, finding similar effects on γH2AX foci formation compared to mouse HSPCs (Supplementary Figure S1C-S1E). Since ADAR1p150 is encoded by an inducible transcript of ADAR1 gene and is predominantly cytosolic (45), we hypothesized that ADAR1’s genome-protective effects are attributable to its constitutively expressed nuclear form, ADAR1p110. Indeed, ADAR1p150 overexpression was less effective than ADAR1p110 in suppressing γH2AX accumulation and DNA damage in ADAR1-depleted cells expressing siRNA against the ADAR1 5′UTR (Figure 1E–H). To investigate whether the enzymatic activity of ADAR1p110 (hereinafter referred to as ADAR1 unless otherwise specified) is involved in this process, we generated HeLa cells stably expressing wild type ADAR1 (ADAR1/Wt), its RNA-editing defective variants G712R and H615Q/E617A (36,46), or a mutant devoid of dsRNA binding (EAXXA) (38,47). All but not ADAR1/EAXXA significantly reduced the level of DNA damage in ADAR1-depleted cells (Figure 1E–H). Interestingly, ADAR1 lacking its catalytic domain (ADAR1/ΔCD) behaved similarly to ADAR1/Wt (Figure 1E–H). Furthermore, co-depletion of ADAR1 and MDA5 or PKR, which senses unedited dsRNA in ADAR1-deficient cells, could not neutralize ADAR1 ablation-induced effects (Figure 1I). These data raise the possibility that the dsRNA-binding domain, but not the catalytic activity, of nuclear ADAR1 is indispensable for genome integrity.
ADAR1 is physically associated with TOPBP1
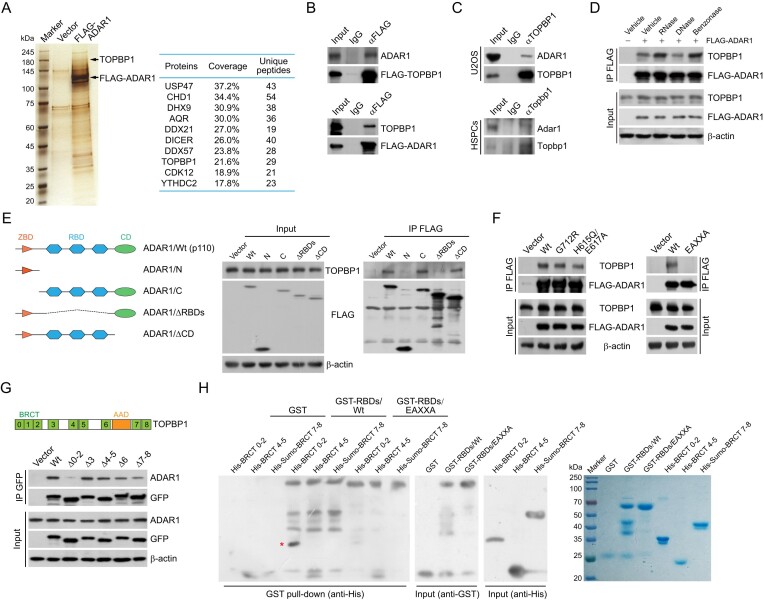
To understand how ADAR1 controls genome stability, we used affinity purification and mass spectrometry to identify the interactome of ADAR1 with cellular extracts from HeLa cells stably expressing FLAG-ADAR1. TOPBP1, a well-defined activator of ATR (7), was one of the top candidates among proteins that copurified with FLAG-ADAR1 (Figure 2A and Supplementary Table S5). Since ATR signaling is essential for the maintenance of HSPC genome integrity and survival during hematopoietic progenitor expansion (48,49), we next performed co-immunoprecipitation (co-IP) to examine the interaction of ADAR1 with TOPBP1. We found that ADAR1 could be efficiently immunoprecipitated by FLAG-tagged TOPBP1 in HeLa cells, and vice versa (Figure 2B). Moreover, we confirmed that endogenous ADAR1 and TOPBP1 form a protein complex in both U2OS cells and HSPCs (Figure 2C). Importantly, DNase or intercalating agent treatment did not change ADAR1–TOPBP1 binding, while benzonase, a pan nuclease that digests DNA and RNA, and RNase markedly enhanced the interaction (Figures 2D and S2A). These data suggest that the TOPBP1–ADAR1 complex is unlikely bridged by nucleic acids, while TOPBP1 may competes with RNA moieties for ADAR1 binding.
Figure 2.
ADAR1 is physically associated with TOPBP1. (A) Analysis of FLAG-ADAR1-associated proteins. Differential protein bands of the immunoprecipitates from HeLa cell extracts were retrieved from silver-stained SDS-PAGE, digested with trypsin, and analyzed by mass spectrometry. The parameters of the representatively top 10 candidates are shown. (B) Immunoprecipitation (IP) followed by immunoblotting (IB) with cellular extracts from HeLa cells expressing FLAG-TOPBP1 or FLAG-ADAR1. (C) Co-IP analysis of the interaction between TOPBP1 and ADAR1 with cellular extracts from U2OS cells and mouse HSPCs. (D) Co-IP analysis of the association of TOPBP1 with FLAG-ADAR1 in the absence or presence of different nucleases as indicated. (E, F) Co-IP analysis of the association of TOPBP1 with the indicated FLAG-tagged ADAR1 mutants in HeLa cells. A schematic of ADAR1 domains and its deletion mutants is shown in (E). (G) Co-IP analysis of the interaction of ADAR1 with GFP-tagged TOPBP1 deletion mutants. A schematic of TOPBP1 domains is shown. BRCT, BRCA1 C-terminal; AAD, ATR activation domain. (H) GST pull-down assays with recombinant ADAR1 RNA-binding domains (GST-RBDs/Wt or GST-RBDs/EAXXA) and TOPBP1 BRCT domains (His-BRCT 0–2, His-BRCT 4–5 and His-Sumo-BRCT 7–8). High salt and heparin column was used to remove contaminating RNAs for RBD purification. The recombinant proteins were examined by Coomassie brilliant blue staining and immunoblotting. The pulled-down protein is marked by a red asterisk.
To further understand the interaction details between ADAR1 and TOPBP1, we generated FLAG-tagged domain deletion mutants of ADAR1. Co-IP showed that the three RBDs are required for ADAR1 binding to TOPBP1 (Figure 2E). Interestingly, the dsRNA-binding defective mutant ADAR1/EAXXA was unable to bind TOPBP1 (Figure 2F), and KKXXK motif of the third RBD contributes most to the interaction (Supplementary Figure S2B). Immunoprecipitation with domain deletion mutants of TOPBP1 revealed that BRCT 0–2 is responsible for TOPBP1 association with ADAR1 (Figure 2G). ADAR1 phosphorylation is apparently not required for this interaction, as λ-phosphatase treatment did not abolish this binding (Supplementary Figure S2C). We next purified GST-tagged RBDs of ADAR1 and His-tagged BRCTs of TOPBP1 from bacteria. GST pull-downs with these recombinant proteins demonstrated that BRCT 0–2 was specifically responsible for TOPBP1 interaction with ADAR1’s RBDs, while EAXXA mutation nearly abolished this binding (Figure 2H). These results suggest that ADAR1 and TOPBP1 interact via their RBDs and BRCT 0–2 domains, respectively, while RNAs do not contribute to the formation of ADAR1–TOPBP1 complex.
ADAR1 is an essential regulator of ATR activation
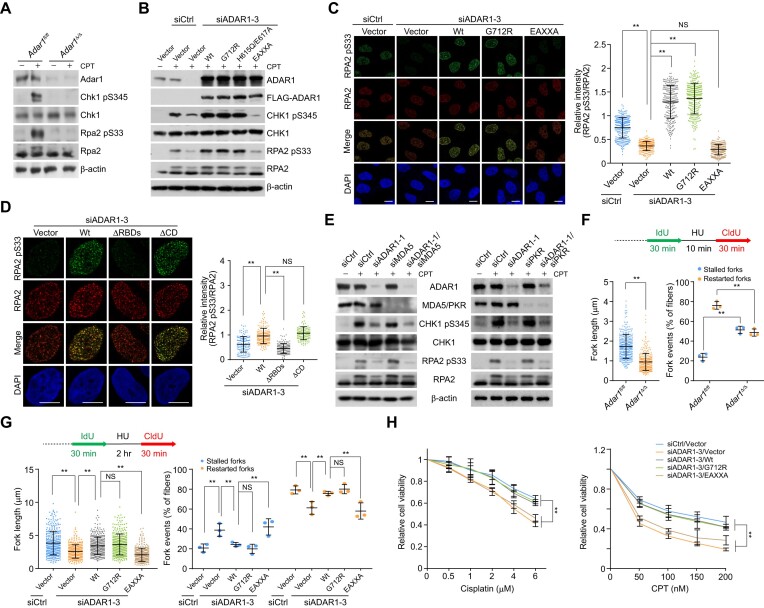
To assess whether ADAR1 plays a role in ATR activation, we cultured HSPCs in the presence of the topoisomerase inhibitor camptothecin (CPT) to arrest replication forks. Immunoblotting indicated that Adar1 knockout significantly decreased the phosphorylation of CHK1 S345 and RPA2 S33, which are canonical substrates of ATR (6) (Figure 3A). Similar results were observed in HeLa cells transfected with ADAR1 siRNAs (Supplementary Figure S3A). The intensity of RPA2 S33 foci in ADAR1-depleted cells was also significantly reduced, while RPA2 foci formation was essentially unchanged (Supplementary Figure S3B). Furthermore, ATR kinase activity toward RPA2 S33 was markedly impaired when nuclear extracts from ADAR1-depleted cells were incubated with a replication fork analogue carrying a hairpin-like ssDNA-dsDNA with 5′ junction, a compatible structure for TOPBP1 loading (50) (Supplementary Figure S3C). These results suggest that ADAR1 is required for ATR activation in the replication stress response.
Figure 3.
ADAR1 is an essential regulator of ATR activation. (A) Immunoblotting analysis of ATR kinase activity in Adar1-knockout HSPCs. Cells were treated with camptothecin (CPT; 1 μM) for 1 h before collection, and the levels of the indicated proteins and drug-induced phosphorylation events were examined. (B) Immunoblotting analysis of ATR kinase activity in HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants. Cells were treated with CPT (1 μM) for 1 h before collection. (C, D) Immunostaining and confocal microscopy analysis of RPA2 pS33 and RPA2 foci formation in HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants (n > 200 for C; n > 130 for D). Cells were treated with CPT for 1 h followed by pre-extraction and fixation. Scale bars, 10 μm. (E) Analysis of ATR kinase activity by immunoblotting with cellular extracts from HeLa cells expressing the indicated siRNAs. Cells were treated with CPT for 1 h before collection. (F) Analysis of the tract lengths of restarted replication forks and the percentage of stalled or restarted forks by DNA fiber assay in control or Adar1-knockout HSPCs. HSPCs were treated with hydroxy urea (HU; 2 mM) for 10 min between IdU and CldU labeling to arrest replication forks. The tract lengths of restarted replication forks (CldU only) and the percentage of stalled (IdU only) or restarted (IdU-CldU) forks were quantified (n > 320). (G) Analysis of tract lengths of restarted replication forks and the percentage of stalled or restarted forks by DNA fiber assay in HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants. Cells were treated with HU (2 mM) for 2 h between the labeling intervals (n > 300). (H) Survival analysis of cisplatin- or CPT-treated HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants. Data are mean ± SDs for (C, D) and (F–H) from biological triplicate experiments. **P < 0.01; NS, not significant; Kruskal-Wallis test for (C), (D) and (G, left); Mann–Whitney test for (F, left); one-way ANOVA for (F, right and G, right); and two-way ANOVA for (H).
To determine whether ATR activation relies on ADAR1-TOPBP1 binding or ADAR1’s enzymatic activity, we examined CHK1 and RPA2 phosphorylation in HeLa cells expressing ADAR1 variants and siRNA against the 5′UTR of ADAR1. Immunoblotting and immunofluorescence revealed that only the EAXXA mutant was unable to revert ATR’s kinase activity and restore RPA2 pS33 foci formation in ADAR1-depleted cells (Figure 3B, C). Consistently, ADAR1 lacking the C-terminal catalytic domain (ΔCD), but not the variant devoid of RBDs (ΔRBDs), also stimulated ATR activity (Figure 3D). Co-depletion of ADAR1 and MDA5 or PKR caused effects similar to ADAR1 knockdown (Figure 3E). These data indicate that ADAR1-mediated ATR activation requires ADAR1’s RBDs, but not its RNA editing activity.
We then investigated whether ADAR1 regulates the recovery of DNA synthesis after transient replication block, which requires intact ATR kinase activity (51). To this end, we analyzed the effect of ADAR1 loss on the status of replication forks via DNA fiber assays at single-molecule resolution. Newly synthesized DNA was labeled with 5-iodo-2′-deoxyuridine (IdU), treated with hydroxyurea (HU), an inhibitor of the ribonucleotide reductase, and then labeled with 5-chloro-2′-deoxyuridine (CldU) after removing the inhibitor. Individual replication tracts were visualized by immunofluorescence microscopy. ADAR1 knockout or knockdown significantly reduced replication strand lengths (CldU-labeled replication tracts), increased the fraction of stalled replication forks (IdU-only tracts), and reduced replication fork restart (IdU-CldU tracts) (Figures 3F and S3D), indicating that ADAR1 is required for appropriate fork restart and progression in the replication stress response. Furthermore, overexpression of wt or G712R ADAR1 efficiently compensated for the replication defects induced by ADAR1 depletion, while EAXXA did not (Figure 3G). These observations suggest that RBDs’ integrity is necessary for DNA synthesis recovery and replication fidelity. Cisplatin or CPT treatment consistently reduced the viability of ADAR1-depleted cells, and rescue experiments showed that these defects could not be overcome by ADAR1/EAXXA (Figure 3H). Collectively, we propose that the genome-protective role of ADAR1 depends on its functional RBDs rather than A-to-I editing activity.
ADAR1 controls the recruitment of TOPBP1 to replication stress sites
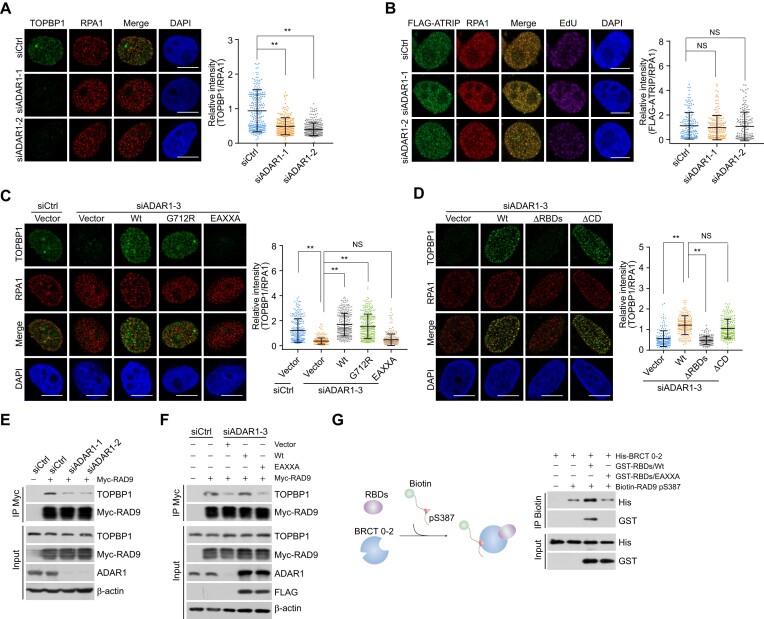
To understand how ADAR1 regulates ATR activation, we examined whether ADAR1 is required for the recruitment of TOPBP1 to replication stress sites. First, the distribution of TOPBP1, reflected by its foci formation at perturbed replication forks was monitored in cells expressing ADAR1 siRNAs. The recruitment of TOPBP1, but not ATRIP, was significantly disrupted by ADAR1 knockdown in CPT-treated cells (Figure 4A, B). By contrast, ADAR1 depletion did not affect TOPBP1 foci formation at nuclease FokI-generated double-strand breaks (DSBs) around LacO sites (52,53) (Supplementary Figure S4A) or the chromatin-binding affinity of TOPBP1 in native cells (Supplementary Figure S4B, C). Next, we examined whether ADAR1-TOPBP1 binding is involved in TOPBP1 loading onto replication stress sites. We found that ADAR1/EAXXA- and ADAR1/ΔRBDs-expressing cells exhibited severe TOPBP1 loading failure (Figure 4C, D). Enhanced recruitment of TOPBP1 triggered by ADAR1 overexpression provides a possible explanation for the increased ATR activity observed in ADAR1/Wt- and ADAR1/G712R-expressing cells (Figure 3B, C). Collectively, these results indicate that ADAR1 directs TOPBP1 engagement at damaged replication forks.
Figure 4.
ADAR1 controls the recruitment of TOPBP1 to replication stress sites. (A) Immunostaining and confocal microscopy analysis of TOPBP1 and RPA1 foci formation in ADAR1-depleted U2OS cells. Cells were treated with CPT for 1 h followed by pre-extraction and fixation. The intensity of TOPBP1 foci was quantified and normalized to that of RPA1 foci (n > 200). Scale bars, 10 μm. (B) Immunostaining and confocal microscopy analysis of the recruitment of FLAG- ATRIP at replication stress sites in HeLa cells expressing siRNAs against ADAR1. Cells were treated with CPT for 1 h followed by pre-extraction and fixation. The intensity of ATRIP foci was quantified and normalized to that in control cells (n > 100). Scale bars, 10 μm. (C, D) Immunostaining and confocal microscopy analysis of TOPBP1 and RPA1 foci formation in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated stably integrated ADAR1 variants. Cells were treated with CPT for 1 h before collection. The intensity of TOPBP1 foci was quantified and normalized to that of RPA1 foci (n > 200 for C; n > 110 for D). Scale bars, 10 μm. (E) Co-IP analysis of the interaction between TOPBP1 and RAD9 in control or ADAR1-knockdown HeLa cells. (F) Co-IP analysis of the interaction between TOPBP1 and RAD9 in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated ADAR1 variants. (G) Pull-down assays with biotinylated RAD9 pS387 peptides and the indicated recombinant proteins. ADAR1 RBDs/Wt or RBDs/EAXXA was incubated with TOPBP1 BRCT 0–2 followed by addition of the biotinylated RAD9 pS387 peptides. The protein complexes were then captured with streptavidin beads for immunoblotting. Data are mean ± SDs for (A–D) from biological triplicate experiments. **P < 0.01; NS, not significant; Kruskal–Wallis test for (A–D).
Since phosphorylated RAD9 is a primary docking signal for TOPBP1 at replication stress sites, we hypothesized that ADAR1 controls TOPBP1 deposition through its impact on RAD9-TOPBP1 binding. Indeed, we found that ADAR1 depletion weakened the interaction of RAD9 with TOPBP1 (Figure 4E). This was reversible by overexpression of ADAR1/Wt, but not ADAR1/EAXXA (Figure 4F). Next, in vitro pull-down experiments with recombinant proteins revealed that wt RBDs (GST-RBDs/Wt) enhanced TOPBP1 BRCT 0–2 binding to the RAD9 pS387 peptide, while mutated RBDs (GST-RBDs/EAXXA) failed to do so (Figure 4G). Considering that some A-to-I editing events can also impact protein expression (45), we re-analyzed the ADAR1 editome in human and mouse cells with GEO datasets GSE99249 and GSE142216 (36,54). Although gene ontology analysis identified ADAR1 editing sites in a cluster of genes related to DNA repair and replication (Supplementary Figure S4D and Supplementary Table S6), the majority of proteins encoded by these genes were essentially unchanged in ADAR1-deficient cells (Supplementary Figure S4E). These results further reduce the possibility that ADAR1-regulated replication stress response is a secondary effect of RNA recoding. We propose instead that ADAR1 may orchestrate TOPBP1 binding to phosphorylated RAD9, thus facilitating TOPBP1 loading on chromatin at damaged replication forks.
ADAR1 protects the genome against R-loops
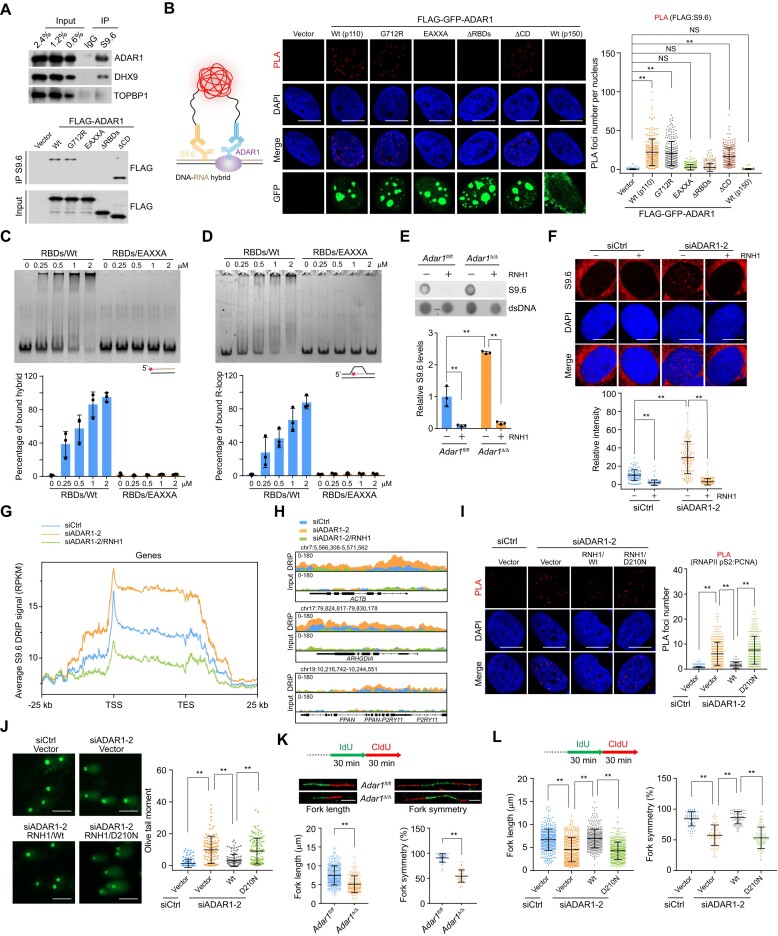
The requirement of ADAR1 in ATR activation may protect against endogenous replicative stress, but this does not fully explain the phenotypes observed in ADAR1-deficient cells. Given that ADAR1 loss-induced DNA damage predominantly occurs in the S phase of the cell cycle and that R-loops are a major source of replication stress and genome instability (13), we hypothesized that ADAR1 is involved in R-loop homeostasis. To test this, we performed immunoprecipitation with fragmented chromatins (∼200 bp) followed by immunoblotting, finding that ADAR1 and the R-loop regulator DHX9 (16,24,55) could be efficiently immunoprecipitated by the S9.6 antibody against DNA–RNA hybrid structures (Figure 5A). Unlike ADAR1, TOPBP1 was not efficiently pulled-down by fragmented DNA–RNA hybrids (Figure 5A), suggesting that TOPBP1 is not involved in R-loop formation as previously reported (32). Furthermore, we demonstrated that ADAR1’s RBDs and key dsRNA-binding residues are responsible for its interaction with DNA–RNA hybrids (Figure 5A). We observed similar results using proximity ligation assays (PLAs), which allow in situ detection of protein-protein or protein-nucleic acid interactions (56), to monitor the in vivo association of ADAR1 variants with R-loop structures (Figure 5B). In agreement with the binding theme for dsRNA (47), the variant bearing deaminase domain while lacking RBDs exhibited quite weaker affinity to DNA–RNA hybrids (Figure 5A-B). Treatment with the RNA polymerase II inhibitor DRB or RNH1/Wt nearly eliminated the PLA signals, further indicating that ADAR1 specifically localizes to transcription-coupled R-loops (Supplementary Figure S5A, B). Meanwhile, we found that the PLA signals between R-loops and FLAG-tagged ADAR1/Wt, ADAR1/G712R, or ADAR1/ΔCD were stronger in EdU-positive cells than those in EdU-negative cells (Supplementary Figure S5C), suggesting that the interaction of R-loops and ADAR1 is not specific to S-phase, but TRCs in actively cycling cells stimulate more R-loop formation and thus ADAR1 binding. To validate the ability of RBDs to recognize DNA–RNA hybrids, we performed electrophoretic mobility shift assays (EMSAs) with annealed ssDNA and ssRNA molecules and recombinant RBDs/Wt or its EAXXA-mutant counterpart. RBDs/Wt shifted the DNA–RNA hybrids in a dose-dependent manner, while RBDs/EAXXA did not (Figure 5C). Similar results were observed when a reconstituted R-loop mimetic structure with displaced ssDNA was used as the substrate (Figure 5D). Consistent with the functional requirement of RBDs in telomeric R-loop resolution (41), these data strongly suggest that ADAR1 can directly interact with R-loops via its RBDs.
Figure 5.
ADAR1 protects the genome against R-loops. (A) IP analysis of R-loop-associated factors, using S9.6 antibody followed by IB. (B) PLA analysis of the localization of FLAG-GFP-tagged ADAR1 variants on R-loops in HeLa cells. The PLA signals of FLAG-ADAR1 and S9.6 were quantified (n > 210). Scale bars, 10 μm. (C) Evaluation of the binding of RBDs to DNA–RNA hybrid. EMSAs were performed with 50 nM Cy3-labeled DNA–RNA hybrid (90 nt) and an increasing amount of recombinant RBDs. The proportion of RBDs-bound DNA–RNA hybrid was quantified. (D) Analysis of the binding of RBDs to R-loop as in (C). (E) Examination of R-loops in control and Adar1-knockout HSPCs via dot blotting. Total DNA were extracted and stained with S9.6 antibody in the presence or absence of recombinant RNH1. The level of R-loops in each sample was quantified and normalized to that of the input DNA. (F) Immunostaining and confocal microscopy analysis of R-loop levels in ADAR1-depleted HeLa cells. Cells were untreated or treated with RNH1 for 4 h before immunostaining with S9.6 antibody. The intensity of nuclear S9.6 signal in each cell was quantified (n > 100). Scale bars, 10 μm. (G) Metaplots of R-loop signals detected by S9.6 DRIP-seq in HeLa cells under the indicated treatments. The color of each line represents the average density of reads for different groups from biological duplicate data. (H) Track examples of DRIP-seq for R-loop alterations around the indicated genes. (I) Analysis of TRCs in HeLa cells expressing ADAR1 siRNA and the indicated stably integrated RNH1 variants. The PLA signal between RNAPII pS2 and PCNA in each cell was quantified (n > 200). Scale bars, 10 μm. (J) Examination of damaged DNA accumulation by alkaline comet assay in HeLa cells expressing ADAR1 siRNA and the indicated stably integrated RNH1 variants (n > 100). Scale bars, 100 μm. (K) DNA fiber assay analysis of the tract lengths and replication fork symmetry in control and Adar1-knockout HSPCs. Fork length (n > 300) and symmetry (n > 55) were determined by measuring CldU tract length. Representative results of altered fork length and symmetry are shown. Scale bars, 5 μm. (L) DNA fiber assay analysis of the tract lengths (n > 390) and replication fork symmetry (n > 55) in HeLa cells expressing ADAR1 siRNA and the indicated stably integrated RNH1 variants. Data are mean ± SDs for (B), (E, F) and (I–L) from biological triplicate experiments. **P < 0.01; NS, not significant; Kruskal–Wallis test for (B), (F), (I–J) and (L), one-way ANOVA for (E) and Mann–Whitney test for (K).
We next sought to verify whether ADAR1 functions in R-loop homeostasis. Dot blotting analysis of total DNA revealed that R-loop levels, reflected by S9.6 signals, were elevated in ADAR1-deficient cells (Figure 5E and S5D). Similar to SRSF1 depletion (26), ADAR1 knockdown significantly increased the S9.6 nuclear immunofluorescence signal (Figure 5F and S5E). To confirm specificity, we removed the S9.6 signals by in vitro treatment with RNH1 (Figure 5E,F). We next examined the R-loop profile along the genome in ADAR1-depleted HeLa cells via DNA–RNA immunoprecipitation (DRIP, here called S9.6 DRIP) coupled with high-throughput sequencing. The results confirmed the predisposition of ADAR1-deficient cells to accumulate DNA–RNA hybrids at the gene body, promoters, and downstream regions, while RNH1 treatment efficiently degraded R-loop-prone regions along the genome (Figure 5G, H). Furthermore, we identified about 1800 genomic sites with higher R-loop content in ADAR1-depleted cells, and the majority of changes (∼82%) occurred within the genic regions (Supplementary Table S7). Based on these data, we propose that ADAR1 functions directly to prevent genome-wide R-loop accumulation.
Co-transcriptional R-loops could aggravate TRCs by creating an additional barrier to replication forks (13,14)—therefore, we assessed whether ADAR1-regulated R-loop homeostasis is also involved in TRC suppression. We first measured the collisions between transcription and replication machineries via PLA with antibodies against the elongating form of RNA polymerase II (RNAPII pS2) and PCNA. ADAR1 depletion significantly augmented PLA signals, which were in turn reduced by CDC7 inhibitor (XL413, which suppresses replication origin firing) and RNAPII inhibitor (DRB, which inhibits Pol II travel) (Supplementary Figure S5F). To test whether R-loops are involved in TRCs, we transfected RNH1 variants into ADAR1-knockdown cells. Only wild type RNH1 (RNH1/Wt), but not its catalytically inactive mutant D210N (RNH1/D210N) (57), could reduce PLA foci of TRCs associated with ADAR1 depletion (Figure 5I). Along with the increased TRC formation observed in ADAR1-knockdown cells (Supplementary Figure S5F), ADAR1 depletion led to more R-loop accumulation compared to ATR inhibition (Supplementary Figure S5G). These results suggest that ADAR1-promoted R-loop clearance is required for TRC suppression.
Finally, we asked whether R-loops are responsible for the increased DNA damage observed in ADAR1-deficient cells. Indeed, overexpression of RNH1/Wt, but not RNH1/D210N, significantly reduced ADAR1 depletion-associated comet tail and γH2AX foci formation (Figures 5J and S5H). Furthermore, in DNA fiber assays, Adar1 knockout or ADAR1 knockdown decreased fiber lengths by ∼35% compared to controls and compromised the symmetry of bidirectional replication forks (Figures 5K and S5I). Consistent with the R-loop dependency of fork elongation control (32,58,59), replication defects including slower fork velocity and asymmetry in ADAR1-deficent cells were significantly ameliorated in RNH1/Wt-expressing cells (Figure 5L). Taken together, these data indicate that ADAR1 plays a critical role in protecting the genome from R-loops. As abnormal R-loop accumulation can lead to TOPBP1-dependent ATR activation (26), we speculate that ADAR1 depletion-associated genome instability is attributable to a synergistic effect from increasing R-loops and decreasing ATR activity.
ADAR1 suppresses R-loop formation in an RNA editing-independent manner
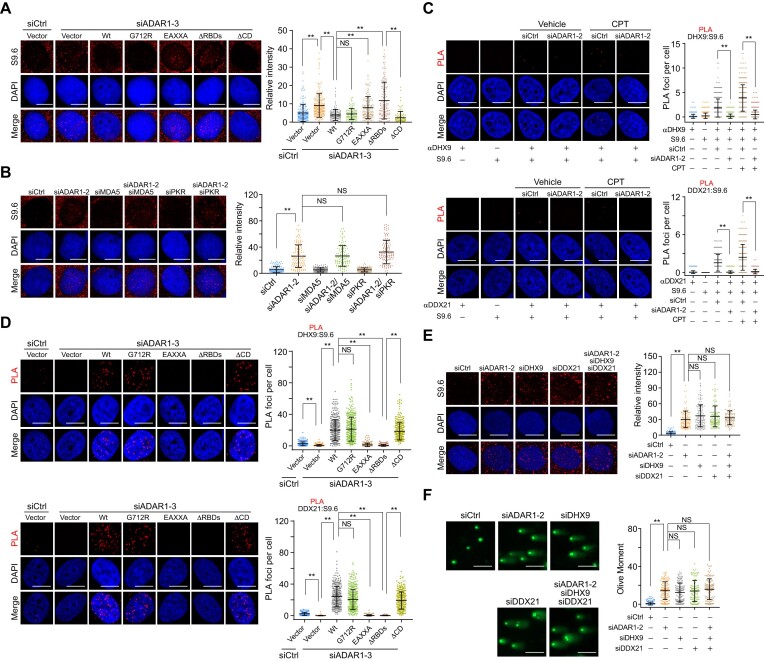
We next aimed to address how ADAR1 protects genomes from R-loop formation. First, we determined the ability of ADAR1 variants to suppress R-loops generated in ADAR1-depleted cells. Consistent with Shiromoto's study (41), DRIP experiments followed by telomeric probe hybridization revealed that ADAR1 depletion resulted in an increased formation of DNA–RNA hybrids at telomeric repeats, and this effect could be rescued by ADAR1/Wt, but not its RNA editing-defective mutant G712R and R-loop binding failure mutant EAXXA (Supplementary Figure S6A, B). By contrast, quantitative PCR analysis of the DRIP products with primers covering the intron 1 region of ACTB revealed that both ADAR1/Wt and ADAR1/G712R, but not ADAR1/EAXXA, were able to suppress R-loop accumulation induced by ADAR1 depletion (Supplementary Figure S6C). Similar to R-loop alterations on ACTB gene, nuclear-wide S9.6 stainings showed that ADAR1/G712R reduced R-loop levels as efficiently as ADAR1/Wt, while ADAR1/EAXXA did not (Figure 6A). This is consistent with the R-loop binding profile of the ADAR1 variants (Figure 5A, B) and their genome protection roles (Figure 1E-G). Remarkably, we found that ADAR1/ΔCD, but not ADAR1/ΔRBDs, could also efficiently suppress R-loop formation (Figure 6A), suggesting that the deaminase activity of ADAR1 is dispensable for global R-loop suppression. In agreement with these findings, co-depletion of ADAR1 and MDA5 or PKR was not able to revert ADAR1 knockdown-associated R-loop formation (Figure 6B). Collectively, these results argue for a non-telomeric R-loop-resolving function of ADAR1 independent of its RNA editing activity but dependent on R-loop recognition.
Figure 6.
ADAR1 suppresses R-loop formation in an RNA editing-independent manner. (A) Immunostaining and confocal microscopy analysis of R-loop levels in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated stably integrated ADAR1 variants. HeLa cells were fixed and stained with S9.6 antibody, and the intensity of nuclear S9.6 signal in individual cells was quantified (n > 100). Scale bars, 10 μm. (B) Examination and quantitative analysis of R-loop levels in HeLa cells expressing the indicated siRNAs (n > 100). Scale bars, 10 μm. (C) Examination and quantitative analysis of the localization of DHX9 and DDX21 on R-loops by PLA in ADAR1-depleted HeLa cells in the presence or absence of CPT (1 μM, 1 h). The PLA signal between R-loops and DHX9 (n > 340) or DDX21 (n > 220) in each cell was quantified. Scale bars, 10 μm. (D) Examination and quantitative analysis of the localization of DHX9 and DDX21 on R-loops by PLA in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated FLAG-tagged ADAR1 variants. The PLA signal between R-loops and DHX9 (n > 210) or DDX21 (n > 200) in each cell was quantified. Scale bars, 10 μm. (E) Immunostaining and confocal microscopy analysis of R-loop levels in HeLa cells expressing the indicated siRNAs. The intensity of nuclear S9.6 signal in individual cells was quantified (n > 110). Scale bars, 10 μm. (F) Examination and quantitative analysis of damaged DNA accumulation by alkaline comet assay in HeLa cells expressing the indicated siRNAs (n > 100). Scale bars, 100 μm. Data are mean ± SDs from biological triplicate experiments. **P < 0.01; NS, not significant; Kruskal–Wallis test.
Given that RNA helicases DHX9 and DDX21, which act as R-loop suppressors by unwinding DNA–RNA hybrids (16,22,23), are potential interactors of ADAR1 (Figure 2A) (60–63), we hypothesized that ADAR1 may rely on these helicases to resolve R-loops. We first examined the recruitment of these helicases at R-loop regions in ADAR1-depleted cells and demonstrated that ADAR1 depletion significantly impaired the PLA signals between DNA–RNA hybrid and DHX9 or DDX21, regardless of the presence or absence of exogenous replication stress (Figures 6C and S6D, E). The expression of DHX9 and DDX21 was not altered in ADAR1-depleted cells (Supplementary Figure S6F). Overexpression in ADAR1-deficient cells of ADAR1/Wt, ADAR1/G712R or ADAR1/ΔCD, but not of R-loop binding failure mutants ADAR1/EAXXA and ADAR1/ΔRBDs, significantly promoted the relocation of DHX9 and DDX21 to R-loops, albeit to different extents (Figure 6D). The PLA signals in these cells were comparable to those for S9.6 and ADAR1 variants in Figure 5B, supporting the sufficiency and necessity of ADAR1 in recruiting these helicases. When ADAR1 was co-depleted with DHX9 and DDX21, R-loops and DNA damage did not change significantly compared to ADAR1 depletion alone (Figure 6E-F and S6G), implying that ADAR1 and these helicases function in the same pathway to prevent R-loop accumulation. Taken together, these results suggest that ADAR1 resolves R-loops by physically engaging RNA helicases DHX9 and DDX21.
R-loop-induced ADAR1 redistribution favors ATR activation in replication stress response
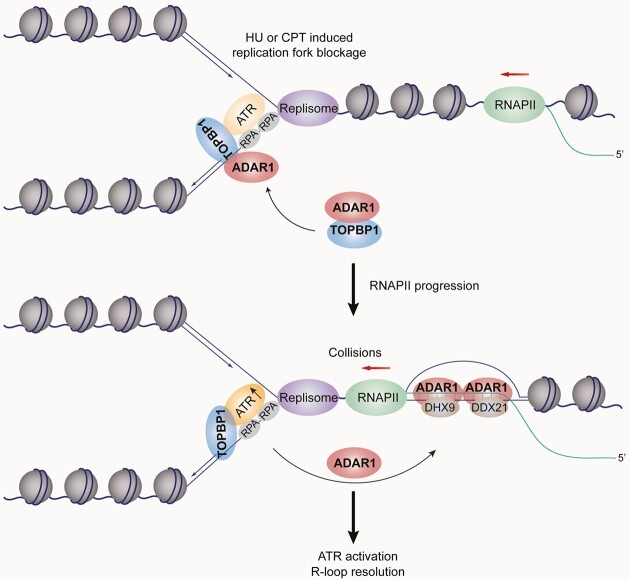
Replication errors or defects may block transcription and generate genome-destabilizing R-loops and TRCs (15–17). For instance, genome-wide mapping of FANCD2 and ATR reveals that a number of stalled replication forks under HU treatment are located close to the pausing RNAPII upstream of transcription start sites (64). Therefore, we asked whether ADAR1-promoted R-loop resolution is functionally linked with ATR activation in the context of replication stress. Co-IP assays demonstrated that either HU or CPT treatment markedly reduced ADAR1–TOPBP1 binding (Figure 7A and S7A) while increasing the interaction between ADAR1 and DHX9 or DDX21 (Figure 7A). Importantly, this alteration could be reverted by either DRB treatment or forced expression of RNH1 (Figure 7A), implying that ADAR1-nucleated protein complexes are dynamically regulated and that R-loops play an important role in controlling ADAR1 redistribution in response to replication stress. To further test this hypothesis, we co-incubated DNA–RNA hybrids with recombinant TOPBP1 BRCT 0–2 and ADAR1/RBDs. GST pull-down analysis (Figure 7B) and co-IP with cellular extracts (Figure 7C) indicated that DNA–RNA hybrid addition impaired TOPBP1 binding to ADAR1 in a dose-dependent manner. Moreover, compared to RNH1/D210N, its wild type counterpart enhanced the colocalization of ADAR1 and TOPBP1 at replication stress sites and weakened ADAR1 and S9.6 PLA signals at R-loops (Figure 7D). In agreement with the immunoprecipitation data in Figure 5A, only a residual amount of TOPBP1 over nuclear background was found to be associated with DNA–RNA hybrids in HU-challenged cells (Figure 7D), confirming that the enhanced interaction of TOPBP1-ADAR1 in RNH1/Wt-expressing cells specifically occurs at blocked replication forks. These observations were further consolidated by in situ analysis of protein interactions at DNA replication forks (SIRF) using proximity ligation coupled with EdU click chemistry in HU-treated cells (65), from which we can see that RNH1/Wt, but not RNH1/D210N, enhanced EdU-ADAR1 interaction, whereas no apparent changes were observed for the PLA signals of EdU-TOPBP1 (Supplementary Figure S7B-S7C). Altogether, these results raise the possibility that replication stress-associated R-loops crosstalk with and actively extract ADAR1 from perturbed replication forks.
Figure 7.
R-loop-induced ADAR1 redistribution favors ATR activation in replication stress response. (A) Co-IP analysis of the interaction of ADAR1 with TOPBP1, DHX9, and DDX21 in HeLa cells stably expressing FLAG-ADAR1 under the indicated treatments. DRB-treated or RNH1-overexpressing HeLa cells were cultured in HU (2 mM) for 4 h before collection. (B) GST pull-down assays with recombinant RBDs and BRCT 0–2 in the presence of an increasing amount of DNA–RNA hybrid or blunt-ended DNA (25 nt) as indicated. The competition effect was quantified. (C) Co-IP analysis of the interaction between ADAR1 and TOPBP1 in the presence of an increasing amount of DNA–RNA hybrid or blunt-ended DNA (25 nt) as indicated. The competition effect was quantified. (D) Examination and quantitative analysis of the distribution of ADAR1 and TOPBP1 on blocked forks or R-loops in HU-treated HeLa cells that stably expressed the indicated RNH1 variants. PLA signal (upper: FLAG-ADAR1 and TOPBP1; middle: FLAG-ADAR1 and S9.6; lower: FLAG-TOPBP1 and S9.6) in each cell was quantified (n > 200). Scale bars, 10 μm. (E) Immunostaining and confocal microscopy analysis of RPA2 pS33 and RPA2 foci formation in HU- and DRB-treated HeLa cells (n > 210). Scale bars, 10 μm. (F) Immunostaining and confocal microscopy analysis of RPA2 pS33 and TOPBP1 foci formation in HeLa cells stably expressing the indicated RNH1 variants (n > 150). Scale bars, 10 μm. (G) Immunoblotting analysis of ATR kinase activity in HeLa cells under the indicated treatments. HeLa cells stably expressing RNH1 variants were transfected with ADAR1/Wt or ADAR1/EAXXA and treated with HU (2 mM) for 4 h before collection. (H) Immunostaining and microscopy analysis of ATR kinase activity in HeLa cells under the indicated treatments. HeLa cells stably expressing ADAR1 were treated with DRB (100 μM) and HU (2 mM) for 4 h before collection (n > 200). Scale bars, 10 μm. (I) A schematic of immunoprecipitation and immunoblotting experiments with nuclear extracts and various types of nucleic acids as indicated. (J) Co-IP analysis of the composition of ADAR1-nucleated protein complexes with nuclear extracts from ADAR1-overexpressing HeLa cells in the presence of the indicated nucleic acids as shown in (I). (K) Examination of ATR kinase activity with nuclear extracts from ADAR1-overexpressing or -knockdown HeLa cells in the presence of the indicated nucleic acids as shown in (I). (L) Examination of ATR kinase activity with nuclear extracts from ADAR1-overexpressing HeLa cells in the presence of reconstituted R-loop. Data are mean ± SDs for (B–F) and (H) from biological triplicate experiments. **P < 0.01; NS, not significant; two-way ANOVA for (B, C), Mann–Whitney test for (D) and (E), and Kruskal–Wallis test for (F) and (H).
To address the biological significance of the redistribution of ADAR1, we examined whether R-loops contribute to ATR activation at perturbed replication forks. Immunostaining revealed that DRB treatment significantly reduced RPA2 pS33 levels in HU- or CPT-challenged cells (Figures 7E and S7D). Meanwhile, overexpression of RNH1/Wt, but not RNH1/D210N, markedly decreased RPA2 pS33 levels (Figures 7F and S7E) but did not significantly affect RPA2 or TOPBP1 loading at replication stress sites (Figures 7E, F and S7D, E). These data imply that R-loops promote ATR activation at these sites. Since ADAR1 dissociates from TOPBP1 at perturbed replication forks due to competitive binding by R-loops, we proposed that the re-localization of ADAR1 helps TOPBP1 reduce ADAR1’s steric hindrance that may restrict ATR activity. Indeed, we found that overexpression of ADAR1/Wt, but not ADAR1/EAXXA, increased ATR activity in RNH1/D210N expressing cells, while this effect was not observed in RNH1/Wt-expressing cells (Figure 7G). This raises the possibility that the sustained ADAR1-TOPBP1 interaction in the absence of R-loops creates a less permissive microenvironment for ATR activation. Similarly, the elevated ATR activity evoked by ADAR1 was not observed in DRB-treated cells, though these cells still exhibited stronger TOPBP1 loading (Figure 7H). The behavior of ADAR1 in cells bearing higher level of R-loops could be due to enhanced recruitment of TOPBP1 at replication stress sites (Figure 7H) and, synergistically, an active redistribution of ADAR1.
To further corroborate these findings, we incubated a blocked replication fork analogue with nuclear extracts, followed by addition of excess DNA–RNA hybrid (Figure 7I). Although this in vitro system could not truly reflect the regulatory architecture of R-loop-coupled replication forks, it enabled us to monitor the molecular events upon replication stress during which R-loops emerge after fork blockage or arise at fork-distal chromatins even from different chromosomes. Compared to replication fork alone, the presence of DNA–RNA hybrid led to a reformation of ADAR1-nucleated complexes (Figure 7J), similar to the observations in Figure 7A, and stimulated ATR activity to a greater extent (Figure 7K), while ADAR1 depletion decreased ATR activity regardless of the presence or absence of DNA–RNA hybrid (Figure 7K). ADAR1’s ATR-stimulating activity was also detected in the context of a reconstituted replication fork along with a head-on R-loop, implying that R-loop could act in cis to facilitate ADAR1 redistribution and subsequent ATR activation (Figure 7L). Taken together, we propose that, although ADAR1 is initially required for TOPBP1 loading, replication stress-associated R-loops, derived from a non-negligible proportion of fork-proximal ones or fork-distal ones on actively-transcribed chromatins, concomitantly promote ADAR1 displacement from perturbed replication forks to switch TOPBP1 from a less permissive intermediate state to a competent conformation. This activates ATR and relocates more ADAR1 to R-loops for resolution by recruiting DHX9 and DDX21 (Graphical Abstract).
Discussion
ATR kinase is a master regulator of the replication stress response, and its activation requires TOPBP1 loading to RPA-coated ssDNA (6,66,67). Here, we found that ADAR1 interacts with TOPBP1 and facilitates its binding to RAD9, although the mechanisms underlying ADAR1’s control of TOPBP1 engagement at perturbed replication forks remain unclear. Unlike most of the known interactors of TOPBP1 (52,68–71), ADAR1 phosphorylation appears to be dispensable for its binding to TOPBP1 BRCT 0–2 domains, while R-loops arising from a road block by replication inhibition impairs TOPBP1-ADAR1 complex formation. In this scenario, DNA–RNA hybrid competes with TOPBP1 for ADAR1 binding by occupying the same molecular interface in ADAR1’s RBDs. The functionality of ADAR1 in ATR activation relies on its dynamic complexing with TOPBP1, which can be artificially divided into two stages: first, ADAR1 promotes the loading of TOPBP1 onto damaged replication forks but limits ATR activation to low levels. Later, this restriction is ameliorated by R-loops that coexist with blocked replication forks. Although we have identified critical residues for ADAR1 binding to TOPBP1 or R-loops, structural studies of ADAR1’s interactions with these factors are required to provide a better understanding of its role in sustaining genome stability. It would be interesting to see whether other protein complexes in replication stress response are affected by R-loops, and to what extent, ADAR1 redistribution could stimulate ATR activity.
ADAR1p110 has been reported to edit A:C-mismatched base pairs via its A-to-I editing activity in telomeric DNA–RNA hybrids, the resolution of which depends on RNH2 (41). However, our DRIP-seq analysis showed that ADAR1 controls genome-wide R-loop levels, and S9.6 immunostaining in cells expressing enzyme-dead ADAR1 mutant revealed that ADAR1’s catalytic activity is dispensable for R-loop suppression. This discrepancy may be the result of different R-loop detection methods, siRNA sequences, and ADAR1 expressing constructs. An alternative yet more likely interpretation is that ADAR1 utilizes different molecular machineries to control R-loop formation at a defined genomic region, where the extent of A:C mismatches in DNA–RNA hybrid may guide the mode of ADAR1’s actions toward R-loop clearance. This hypothesis is supported by the evidence that A:C mismatches at the editing site of both RNAs and telomeric DNA–RNA hybrids could more efficiently accommodate substrate recognition and modification of ADAR family proteins (41,72–74). ADAR1 has been identified as a candidate of DNA–RNA hybrid-binding proteins from mass spectrometry-based screenings (75), and RBDs were shown to be essential for its editing capacity toward telomeric DNA–RNA hybrids (41). However, there is still a lack of experimental evidence showing that ADAR1 can directly occupy or sense R-loops. In our study, PLAs with different ADAR1 variants demonstrated that ADAR1’s RBDs are responsible for its accumulation at R-loop regions, and EMSAs with recombinant RBDs and different R-loop-mimetic nucleic acid structures confirmed that the conserved KKXXK motifs among RBDs are essential for R-loop binding. We further found that the ADAR1-bound R-loop serves as a scaffolding platform for RNA helicases DHX9 and DDX21, both of which unwind DNA–RNA hybrids via their ATPase activity (16,22,23). It appears that numerous RNA helicases including aquarius (AQR) and DDX57 may also be associated with ADAR1, but their contributions to ADAR1-directed R-loop removal await in-depth exploration in the future. Nevertheless, this study advances our understanding of how ADAR1 recognizes and resolves non-telomeric R-loops and provides a molecular basis for R-loops triggering ADAR1 redistribution upon replication stress.
Unlike the recently proposed function of ADAR1 in ovarian cancer cells wherein it prevents ATR activation by suppressing R-loop accumulation (76), we could not detect an evident decrease of ATR activity in ADAR1-knockout HSPCs in the absence or presence of replication inhibitor stimulation. In addition to the different cell types and depletion approaches we used, the discrepancy may arise from the fact that the regulatory mechanism of ATR activation in response to replication stress is different from that challenged by R-loops in unstressed cells. By contrast to SRSF1 and AQR that prevent R-loop formation and ATR activation (26), ADAR1 deficiency leads to R-loop accumulation but weakens ATR activity upon replication stress. This unique function of ADAR1 enables us to separate the activity of RBD domains in promoting ATR activation and R-loop resolution.
Aberrant R-loops block the progression of replication machinery and elicit TRCs (13,14). Reciprocally, replication stress or deregulated origin firing could also disturb transcription, thus stimulating DNA–RNA hybrid formation and TRCs (15–17,64). Our study highlights the importance of R-loops in stimulating ATR activity at replication inhibitor-challenged forks and demonstrates an ADAR1-mediated feedback signaling circuitry that safeguards the genome against replication stress-associated TRCs. This argument is supported by the following evidence that (1) either transcription inhibition or RNH1 overexpression overtly reduced ATR activity, and (2) once R-loops were degraded, ADAR1 was trapped at replication stress sites, and ATR was not fully activated by ADAR1-bound TOPBP1. It appears that ADAR1 is a critical intermediate that links R-loops to damaged replication forks at TRCs in the cellular response to replication inhibition. ADAR1 likely prepares TOPBP1 in a poised state, while R-loops behave as an agonist that transpose ADAR1 and thereby free TOPBP1 at replication stress sites. ADAR1 transposition may be controlled by conformational changes of R-loops or perturbed replication forks. This could take place at stalled replication fork-proximal R-loops at TRCs or possibly the spatially close ones emerging on the same or different chromosomes. There are multiple recent reports that R-loops at TRC sites can form behind a stalled replication fork (77,78). It is likely that ADAR1 could be translocated from a stalled fork to the DNA–RNA hybrids forming at lagging strand, while an in-depth study needs to be launched to test this possibility. Beyond the beneficial effects of R-loops on gene expression (79), transcription termination (80), and DSB repair (81–83), our study illustrates their biological function in directly optimizing ATR activity and thus fork stability in replication stress response. Since numerous factors regulate both R-loop suppression and replication fork stability (13,27–29,84,85), it is worth investigating whether and how these factors coordinately regulate replication fidelity and R-loop homeostasis.
It is well-documented that ADAR1 restricts overactivation of dsRNA-sensing pathways by A-to-I editing of dsRNAs to avoid an autoinflammatory response (36,86), and mutations or dysfunction of ADAR1 are often associated with type I interferon-triggered immune disorders (46), neurological diseases (36), and anti-tumor immunity (42). However, RNA editing alone could not explain all of the phenotypes associated with ADAR1 loss (87), suggesting that additional roles for ADAR1 must exist. Recently, A-to-I editing-independent functions have been reported to be responsible for ADAR1-mediated optimization of microRNA processing (38) and suppression of cellular senescence (88) or stress-induced apoptosis (39), and recent studies in mice and Drosophila suggest that these editing-independent roles are conserved (89). These data imply that binding of dsRNA or ADAR1-scaffolded protein complexes could elicit distinctively yet essentially important influences beyond RNA editing. Our study demonstrates that ADAR1’s genome-protective role is decoupled from its dsRNA-editing function but is highly dependent on its temporo-spatially regulated protein complex formation and its transposition from blocked replication forks to R-loops. However, dsRNAs that are bound but not edited by ADAR1 may also contribute to genome surveillance, and this awaits further exploration. Since multiple cellular behaviors including senescence, cell survival, and homeostasis of hematopoiesis are tightly associated with DNA damage response (90), it is likely that ADAR1’s genome-protective activity may be involved in these processes. Given the importance of ADAR1 in controlling genome stability, small molecule inhibitors that abolish the binding of RBDs to R-loops and TOPBP1 may be a promising avenue for new chemotherapeutics.
In summary, our study underscores the importance of ADAR1’s RBD-mediated scaffolding activity, including R-loop recognition and protein complex reassembly, in genome surveillance. We present a proof-of-concept for a beneficial role of R-loops in stimulating ATR activity, thereby helping cells resolve replication defects and suggesting the potential of targeting ADAR1’s DNA–RNA hybrid-binding surface for antitumor interventions.
Supplementary Material
Acknowledgements
We thank Dr Daniel Ackerman of Insight Editing London for editing the manuscript during preparation.
Author contributions: B.Z., Y.L., J.Z., Y.W., S.M., H.C., T.C. and L.S. conceived this project; B.Z., Y.L., J.Z., Y.W., C.L., L.L., Y.Q., J.H., X.Z. and S.M. conducted experiments; B.Z., Y.L., J.Z., Y.W., C.L., T.L., C.Z., L.L., Y.Q., J.H., X.Z. and S.M. acquired data; B.Z., Y.L., J.Z., Y.W., C.L., T.L., C.Z., J.H., X.Z., S.M., H.C., T.C. and L.S. analyzed data; J.Y., J.H., J.Y., J.L. and Z.Y. provided technical supports and research resources; B.Z., Y.L., S.M., H.C., T.C. and L.S. wrote the manuscript.
Contributor Information
Biao Zhang, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Tianjin Institutes of Health Science, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin 300020, China.
Yi Li, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Jieyou Zhang, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Yuejiao Wang, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Can Liang, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Ting Lu, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Tianjin Institutes of Health Science, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin 300020, China.
Chunyong Zhang, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Ling Liu, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Yan Qin, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Jiahuan He, Tianjin Institutes of Health Science, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin 300020, China; State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & School of Basic Medicine, Peking Union Medical College, 100006, Beijing, China.
Xiangnan Zhao, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Tianjin Institutes of Health Science, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin 300020, China.
Jia Yu, Tianjin Institutes of Health Science, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin 300020, China; State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & School of Basic Medicine, Peking Union Medical College, 100006, Beijing, China.
Jihui Hao, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Jie Yang, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Mulin Jun Li, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Zhi Yao, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Shuai Ma, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Hui Cheng, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Tianjin Institutes of Health Science, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin 300020, China.
Tao Cheng, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Tianjin Institutes of Health Science, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin 300020, China.
Lei Shi, State Key Laboratory of Experimental Hematology, Haihe Laboratory of Cell Ecosystem, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. All unique reagents generated in this study are available from the corresponding authors with a completed Materials Transfer Agreement. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (91) with the dataset identifier PXD038167. The raw data for DRIP-seq in this study can be found at the following DOI: https://doi.org/10.5281/zenodo.8150197, and the processed data can be accessed via the following DOI: https://doi.org/10.5281/zenodo.8159348.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Natural Science Foundation of China [82230101, 81972660, 81722036 to L.S., 82372767, 82003004 to S.M., 81872289 to L.L., 82100134 to B.Z.]; Ministry of Science and Technology of China [2021YFA1100900 to T.C., 2020YFE0203000 to H.C.]; Haihe Laboratory of Cell Ecosystem Innovation Fund [22HHXBSS00021 to L.S., 22HHXBSS00016 to H.C.]; CAMS Initiative for Innovative Medicine [2021-I2M-1-019 to H.C., 2021-I2M-1-040 to T.C.]; CAMS Fundamental Research Funds for Central Research Institutes [3332021093 to. T.C.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
References
- 1. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaillard H., Garcia-Muse T., Aguilera A.. Replication stress and cancer. Nat. Rev. Cancer. 2015; 15:276–289. [DOI] [PubMed] [Google Scholar]
- 3. Flach J., Milyavsky M.. Replication stress in hematopoietic stem cells in mouse and man. Mutat. Res. 2018; 808:74–82. [DOI] [PubMed] [Google Scholar]
- 4. Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C.et al.. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014; 512:198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J.. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013; 155:1088–1103. [DOI] [PubMed] [Google Scholar]
- 6. Saldivar J.C., Cortez D., Cimprich K.A.. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017; 18:622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumagai A., Lee J., Yoo H.Y., Dunphy W.G.. TopBP1 activates the ATR-ATRIP complex. Cell. 2006; 124:943–955. [DOI] [PubMed] [Google Scholar]
- 8. Mordes D.A., Glick G.G., Zhao R., Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008; 22:1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rappas M., Oliver A.W., Pearl L.H.. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res. 2011; 39:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu S., Opiyo S.O., Manthey K., Glanzer J.G., Ashley A.K., Amerin C., Troksa K., Shrivastav M., Nickoloff J.A., Oakley G.G.. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012; 40:10780–10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vassin V.M., Anantha R.W., Sokolova E., Kanner S., Borowiec J.A.. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 2009; 122:4070–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marechal A., Zou L.. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015; 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia-Muse T., Aguilera A.. R Loops: from Physiological to Pathological Roles. Cell. 2019; 179:604–618. [DOI] [PubMed] [Google Scholar]
- 14. Crossley M.P., Bocek M., Cimprich K.A.. R-Loops as cellular regulators and genomic threats. Mol. Cell. 2019; 73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sordet O., Redon C.E., Guirouilh-Barbat J., Smith S., Solier S., Douarre C., Conti C., Nakamura A.J., Das B.B., Nicolas E.et al.. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009; 10:887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cristini A., Groh M., Kristiansen M.S., Gromak N.. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018; 23:1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hamperl S., Bocek M.J., Saldivar J.C., Swigut T., Cimprich K.A.. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell. 2017; 170:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niehrs C., Luke B.. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020; 21:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hegazy Y.A., Fernando C.M., Tran E.J.. The balancing act of R-loop biology: the good, the bad, and the ugly. J. Biol. Chem. 2020; 295:905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lockhart A., Pires V.B., Bento F., Kellner V., Luke-Glaser S., Yakoub G., Ulrich H.D., Luke B.. RNase H1 and H2 are differentially regulated to process RNA-DNA hybrids. Cell Rep. 2019; 29:2890–2900. [DOI] [PubMed] [Google Scholar]
- 21. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 22. Song C., Hotz-Wagenblatt A., Voit R., Grummt I.. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017; 31:1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan W., Al-Hadid Q., Wang Z., Shen L., Cho H., Wu X., Yang Y.. TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. Nucleic Acids Res. 2021; 49:8573–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chakraborty P., Grosse F.. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.). 2011; 10:654–665. [DOI] [PubMed] [Google Scholar]
- 25. Li X., Niu T., Manley J.L.. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007; 13:2108–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matos D.A., Zhang J.M., Ouyang J., Nguyen H.D., Genois M.M., Zou L.. ATR protects the genome against R loops through a MUS81-triggered feedback loop. Mol. Cell. 2020; 77:514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L.et al.. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 2015; 57:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhatia V., Barroso S.I., Garcia-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A.. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 29. Schwab R.A., Nieminuszczy J., Shah F., Langton J., Lopez Martinez D., Liang C.C., Cohn M.A., Gibbons R.J., Deans A.J., Niedzwiedz W.. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell. 2015; 60:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lam F.C., Kong Y.W., Huang Q., Vu Han T.L., Maffa A.D., Kasper E.M., Yaffe M.B.. BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage. Nat. Commun. 2020; 11:4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bayona-Feliu A., Barroso S., Munoz S., Aguilera A.. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet. 2021; 53:1050–1063. [DOI] [PubMed] [Google Scholar]
- 32. Barroso S., Herrera-Moyano E., Munoz S., Garcia-Rubio M., Gomez-Gonzalez B., Aguilera A.. The DNA damage response acts as a safeguard against harmful DNA–RNA hybrids of different origins. EMBO Rep. 2019; 20:e47250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002; 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. George C.X., Samuel C.E.. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Desterro J.M., Keegan L.P., Lafarga M., Berciano M.T., O’Connell M., Carmo-Fonseca M.. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003; 116:1805–1818. [DOI] [PubMed] [Google Scholar]
- 36. Chung H., Calis J.J.A., Wu X., Sun T., Yu Y., Sarbanes S.L., Dao Thi V.L., Shilvock A.R., Hoffmann H.H., Rosenberg B.R.et al.. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018; 172:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakahama T., Kato Y., Shibuya T., Inoue M., Kim J.I., Vongpipatana T., Todo H., Xing Y., Kawahara Y.. Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutieres-syndrome-like encephalopathy. Immunity. 2021; 54:1976–1988. [DOI] [PubMed] [Google Scholar]
- 38. Ota H., Sakurai M., Gupta R., Valente L., Wulff B.E., Ariyoshi K., Iizasa H., Davuluri R.V., Nishikura K.. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013; 153:575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sakurai M., Shiromoto Y., Ota H., Song C., Kossenkov A.V., Wickramasinghe J., Showe L.C., Skordalakes E., Tang H.Y., Speicher D.W.et al.. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017; 24:534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sagredo E.A., Sagredo A.I., Blanco A., Rojas De Santiago P., Rivas S., Assar R., Perez P., Marcelain K., Armisen R.. ADAR1 Transcriptome editing promotes breast cancer progression through the regulation of cell cycle and DNA damage response. Biochim. Biophys. Acta. Mol. Cell Res. 2020; 1867:118716. [DOI] [PubMed] [Google Scholar]
- 41. Shiromoto Y., Sakurai M., Minakuchi M., Ariyoshi K., Nishikura K.. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells. Nat. Commun. 2021; 12:1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ishizuka J.J., Manguso R.T., Cheruiyot C.K., Bi K., Panda A., Iracheta-Vellve A., Miller B.C., Du P.P., Yates K.B., Dubrot J.et al.. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019; 565:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. XuFeng R., Boyer M.J., Shen H., Li Y., Yu H., Gao Y., Yang Q., Wang Q., Cheng T.. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc. Natl. Acad. Sci. USA. 2009; 106:17763–17768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y.. GammaH2AX and cancer. Nat. Rev. Cancer. 2008; 8:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quin J., Sedmik J., Vukic D., Khan A., Keegan L.P., O’Connell M.A. ADAR RNA modifications, the epitranscriptome and innate immunity. Trends Biochem. Sci. 2021; 46:758–771. [DOI] [PubMed] [Google Scholar]
- 46. Rice G.I., Kasher P.R., Forte G.M., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A.et al.. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 2012; 44:1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valente L., Nishikura K.. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 2007; 282:16054–16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao L., Li D., Ma K., Zhang W., Xu T., Fu C., Jing C., Jia X., Wu S., Sun X.et al.. TopBP1 governs hematopoietic stem/progenitor cells survival in zebrafish definitive hematopoiesis. PLoS Genet. 2015; 11:e1005346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schuler F., Afreen S., Manzl C., Hacker G., Erlacher M., Villunger A.. Checkpoint kinase 1 is essential for fetal and adult hematopoiesis. EMBO Rep. 2019; 20:e47026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zou L., Liu D., Elledge S.J.. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13827–13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008; 9:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao J., Tian S., Guo Q., Bao K., Yu G., Wang X., Shen X., Zhang J., Chen J., Yang Y.et al.. A PARylation-phosphorylation cascade promotes TOPBP1 loading and RPA-RAD51 exchange in homologous recombination. Mol. Cell. 2022; 82:2571–2587. [DOI] [PubMed] [Google Scholar]
- 53. Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., Greenberg R.A.. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013; 20:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang F., He J., Liu S., Gao A., Yang L., Sun G., Ding W., Li C.Y., Gou F., He M.et al.. A comprehensive RNA editome reveals that edited Azin1 partners with DDX1 to enable hematopoietic stem cell differentiation. Blood. 2021; 138:1939–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim S., Kang N., Park S.H., Wells J., Hwang T., Ryu E., Kim B.G., Hwang S., Kim S.J., Kang S.et al.. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020; 48:7218–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S.M., Ostman A., Landegren U.. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002; 20:473–477. [DOI] [PubMed] [Google Scholar]
- 57. Chen L., Chen J.Y., Zhang X., Gu Y., Xiao R., Shao C., Tang P., Qian H., Luo D., Li H.et al.. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell. 2017; 68:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saxena S., Zou L.. Hallmarks of DNA replication stress. Mol. Cell. 2022; 82:2298–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gan W., Guan Z., Liu J., Gui T., Shen K., Manley J.L., Li X.. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011; 25:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cho N.H., Cheveralls K.C., Brunner A.D., Kim K., Michaelis A.C., Raghavan P., Kobayashi H., Savy L., Li J.Y., Canaj H.et al.. OpenCell: endogenous tagging for the cartography of human cellular organization. Science. 2022; 375:eabi6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Havugimana P.C., Goel R.K., Phanse S., Youssef A., Padhorny D., Kotelnikov S., Kozakov D., Emili A.. Scalable multiplex co-fractionation/mass spectrometry platform for accelerated protein interactome discovery. Nat. Commun. 2022; 13:4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chu L., Su M.Y., Maggi L.B. Jr, Lu L., Mullins C., Crosby S., Huang G., Chng W.J., Vij R., Tomasson M.H. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Invest. 2012; 122:2793–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hong H., An O., Chan T.H.M., Ng V.H.E., Kwok H.S., Lin J.S., Qi L., Han J., Tay D.J.T., Tang S.J.et al.. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res. 2018; 46:7953–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Solvie D., Baluapuri A., Uhl L., Fleischhauer D., Endres T., Papadopoulos D., Aziba A., Gaballa A., Mikicic I., Isaakova E.et al.. MYC multimers shield stalled replication forks from RNA polymerase. Nature. 2022; 612:148–155. [DOI] [PubMed] [Google Scholar]
- 65. Roy S., Luzwick J.W., Schlacher K.. SIRF: quantitative in situ analysis of protein interactions at DNA replication forks. J. Cell Biol. 2018; 217:1521–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yazinski S.A., Zou L.. Functions, regulation, and therapeutic implications of the ATR checkpoint pathway. Annu. Rev. Genet. 2016; 50:155–173. [DOI] [PubMed] [Google Scholar]
- 67. Lecona E., Fernandez-Capetillo O.. Targeting ATR in cancer. Nat. Rev. Cancer. 2018; 18:586–595. [DOI] [PubMed] [Google Scholar]
- 68. Day M., Rappas M., Ptasinska K., Boos D., Oliver A.W., Pearl L.H.. BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands. eLife. 2018; 7:e39979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Leimbacher P.A., Jones S.E., Shorrocks A.K., de Marco Zompit M., Day M., Blaauwendraad J., Bundschuh D., Bonham S., Fischer R., Fink D.et al.. MDC1 interacts with TOPBP1 to maintain chromosomal stability during mitosis. Mol. Cell. 2019; 74:571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Blackford A.N., Nieminuszczy J., Schwab R.A., Galanty Y., Jackson S.P., Niedzwiedz W.. TopBP1 interacts with BLM to maintain genome stability but is dispensable for preventing BLM degradation. Mol. Cell. 2015; 57:1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gong Z., Kim J.E., Leung C.C., Glover J.N., Chen J.. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell. 2010; 37:438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vogel P., Schneider M.F., Wettengel J., Stafforst T.. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew. Chem. Int. Ed Engl. 2014; 53:6267–6271. [DOI] [PubMed] [Google Scholar]
- 73. Matthews M.M., Thomas J.M., Zheng Y., Tran K., Phelps K.J., Scott A.I., Havel J., Fisher A.J., Beal P.A.. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 2016; 23:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kuttan A., Bass B.L.. Mechanistic insights into editing-site specificity of ADARs. Proc. Natl. Acad. Sci. USA. 2012; 109:E3295–E3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang I.X., Grunseich C., Fox J., Burdick J., Zhu Z., Ravazian N., Hafner M., Cheung V.G.. Human proteins that interact with RNA/DNA hybrids. Genome Res. 2018; 28:1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cui H., Yi Q., Tian M., Yang H.T., Liang Y., Huang J., Zeng Q., Sun W., Han J., Guo J.et al.. ADAR1 Prevents R-loop Accumulation-Driven ATR Pathway Activation in Ovarian Cancer. J. Cancer. 2022; 13:2397–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Svikovic S., Crisp A., Tan-Wong S.M., Guilliam T.A., Doherty A.J., Proudfoot N.J., Guilbaud G., Sale J.E.. R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J. 2019; 38:e99793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stoy H., Zwicky K., Kuster D., Lang K.S., Krietsch J., Crossley M.P., Schmid J.A., Cimprich K.A., Merrikh H., Lopes M.. Direct visualization of transcription-replication conflicts reveals post-replicative DNA:RNA hybrids. Nat. Struct. Mol. Biol. 2023; 30:348–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Grunseich C., Wang I.X., Watts J.A., Burdick J.T., Guber R.D., Zhu Z., Bruzel A., Lanman T., Chen K., Schindler A.B.et al.. Senataxin mutation reveals how R-loops promote transcription by blocking DNA methylation at gene promoters. Mol. Cell. 2018; 69:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhao D.Y., Gish G., Braunschweig U., Li Y., Ni Z., Schmitges F.W., Zhong G., Liu K., Li W., Moffat J.et al.. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016; 529:48–53. [DOI] [PubMed] [Google Scholar]
- 81. Ouyang J., Yadav T., Zhang J.M., Yang H., Rheinbay E., Guo H., Haber D.A., Lan L., Zou L.. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature. 2021; 594:283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yasuhara T., Kato R., Hagiwara Y., Shiotani B., Yamauchi M., Nakada S., Shibata A., Miyagawa K.. Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell. 2018; 175:558–570. [DOI] [PubMed] [Google Scholar]
- 83. Lu W.T., Hawley B.R., Skalka G.L., Baldock R.A., Smith E.M., Bader A.S., Malewicz M., Watts F.Z., Wilczynska A., Bushell M.. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 2018; 9:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Garcia-Rubio M.L., Perez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., Aguilera A.. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015; 11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liang Z., Liang F., Teng Y., Chen X., Liu J., Longerich S., Rao T., Green A.M., Collins N.B., Xiong Y.et al.. Binding of FANCI-FANCD2 complex to RNA and R-loops stimulates robust FANCD2 monoubiquitination. Cell Rep. 2019; 26:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., Walkley C.R.. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015; 349:1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Licht K., Jantsch M.F.. The other face of an editor: ADAR1 functions in editing-independent ways. Bioessays. 2017; 39:1700129. [DOI] [PubMed] [Google Scholar]
- 88. Hao X., Shiromoto Y., Sakurai M., Towers M., Zhang Q., Wu S., Havas A., Wang L., Berger S., Adams P.D.et al.. ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16(INK4a) levels. Nat. Cell Biol. 2022; 24:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Deng P., Khan A., Jacobson D., Sambrani N., McGurk L., Li X., Jayasree A., Hejatko J., Shohat-Ophir G., O’Connell M.A.et al.. Adar RNA editing-dependent and -independent effects are required for brain and innate immune functions in Drosophila. Nat. Commun. 2020; 11:1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F.et al.. iProX: an integrated proteome resource. Nucleic Acids Res. 2019; 47:D1211–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. All unique reagents generated in this study are available from the corresponding authors with a completed Materials Transfer Agreement. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (91) with the dataset identifier PXD038167. The raw data for DRIP-seq in this study can be found at the following DOI: https://doi.org/10.5281/zenodo.8150197, and the processed data can be accessed via the following DOI: https://doi.org/10.5281/zenodo.8159348.