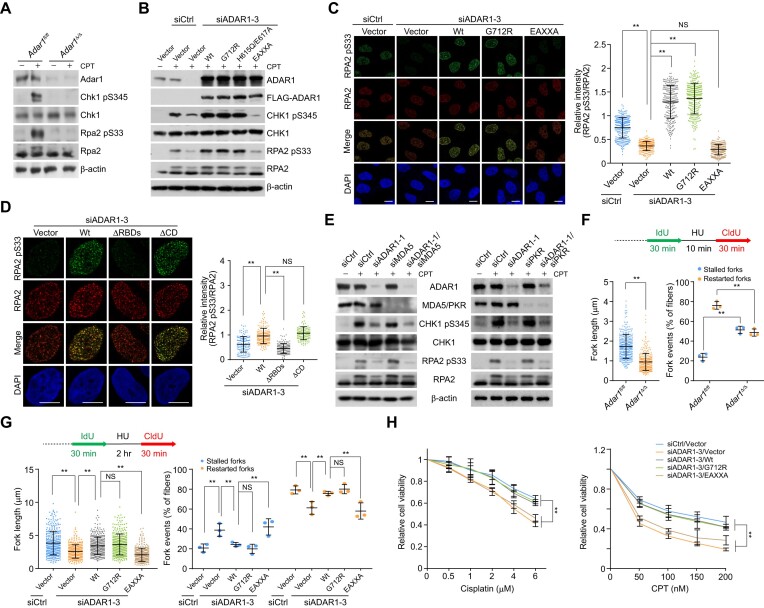
Figure 3.
ADAR1 is an essential regulator of ATR activation. (A) Immunoblotting analysis of ATR kinase activity in Adar1-knockout HSPCs. Cells were treated with camptothecin (CPT; 1 μM) for 1 h before collection, and the levels of the indicated proteins and drug-induced phosphorylation events were examined. (B) Immunoblotting analysis of ATR kinase activity in HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants. Cells were treated with CPT (1 μM) for 1 h before collection. (C, D) Immunostaining and confocal microscopy analysis of RPA2 pS33 and RPA2 foci formation in HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants (n > 200 for C; n > 130 for D). Cells were treated with CPT for 1 h followed by pre-extraction and fixation. Scale bars, 10 μm. (E) Analysis of ATR kinase activity by immunoblotting with cellular extracts from HeLa cells expressing the indicated siRNAs. Cells were treated with CPT for 1 h before collection. (F) Analysis of the tract lengths of restarted replication forks and the percentage of stalled or restarted forks by DNA fiber assay in control or Adar1-knockout HSPCs. HSPCs were treated with hydroxy urea (HU; 2 mM) for 10 min between IdU and CldU labeling to arrest replication forks. The tract lengths of restarted replication forks (CldU only) and the percentage of stalled (IdU only) or restarted (IdU-CldU) forks were quantified (n > 320). (G) Analysis of tract lengths of restarted replication forks and the percentage of stalled or restarted forks by DNA fiber assay in HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants. Cells were treated with HU (2 mM) for 2 h between the labeling intervals (n > 300). (H) Survival analysis of cisplatin- or CPT-treated HeLa cells expressing ADAR1 5′UTR siRNA and the stably integrated ADAR1 variants. Data are mean ± SDs for (C, D) and (F–H) from biological triplicate experiments. **P < 0.01; NS, not significant; Kruskal-Wallis test for (C), (D) and (G, left); Mann–Whitney test for (F, left); one-way ANOVA for (F, right and G, right); and two-way ANOVA for (H).