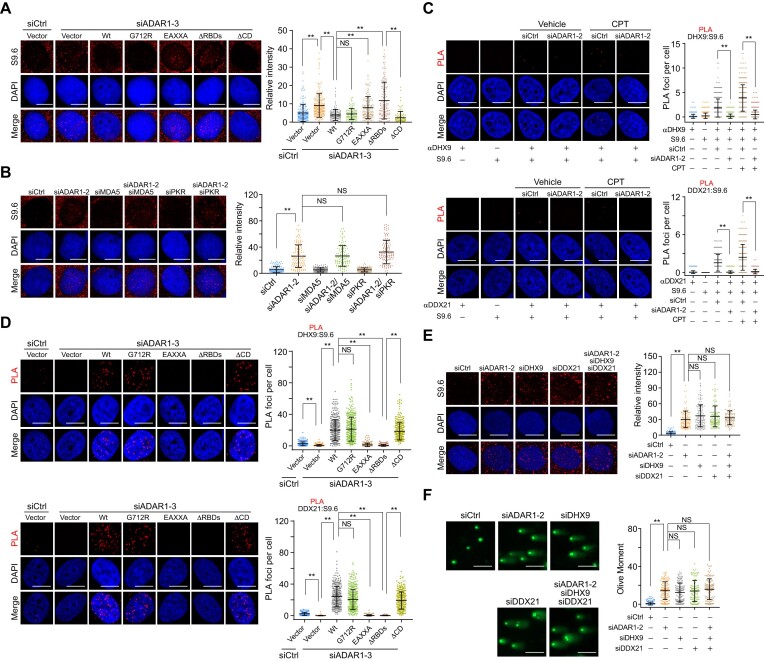
Figure 6.
ADAR1 suppresses R-loop formation in an RNA editing-independent manner. (A) Immunostaining and confocal microscopy analysis of R-loop levels in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated stably integrated ADAR1 variants. HeLa cells were fixed and stained with S9.6 antibody, and the intensity of nuclear S9.6 signal in individual cells was quantified (n > 100). Scale bars, 10 μm. (B) Examination and quantitative analysis of R-loop levels in HeLa cells expressing the indicated siRNAs (n > 100). Scale bars, 10 μm. (C) Examination and quantitative analysis of the localization of DHX9 and DDX21 on R-loops by PLA in ADAR1-depleted HeLa cells in the presence or absence of CPT (1 μM, 1 h). The PLA signal between R-loops and DHX9 (n > 340) or DDX21 (n > 220) in each cell was quantified. Scale bars, 10 μm. (D) Examination and quantitative analysis of the localization of DHX9 and DDX21 on R-loops by PLA in HeLa cells expressing ADAR1 5′UTR siRNA and the indicated FLAG-tagged ADAR1 variants. The PLA signal between R-loops and DHX9 (n > 210) or DDX21 (n > 200) in each cell was quantified. Scale bars, 10 μm. (E) Immunostaining and confocal microscopy analysis of R-loop levels in HeLa cells expressing the indicated siRNAs. The intensity of nuclear S9.6 signal in individual cells was quantified (n > 110). Scale bars, 10 μm. (F) Examination and quantitative analysis of damaged DNA accumulation by alkaline comet assay in HeLa cells expressing the indicated siRNAs (n > 100). Scale bars, 100 μm. Data are mean ± SDs from biological triplicate experiments. **P < 0.01; NS, not significant; Kruskal–Wallis test.