Figure 2.
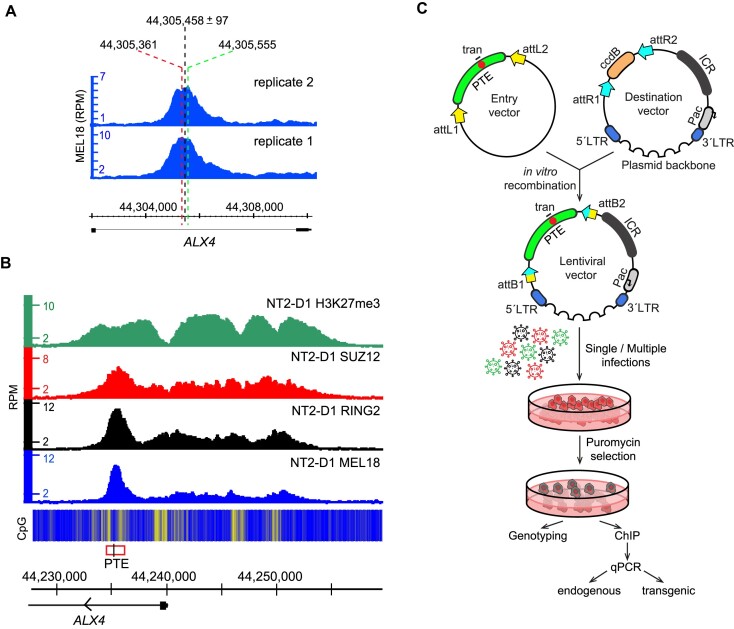
Distinct peaks mark PRC1 tethering elements. (A) Assigning genomic positions of candidate PTEs. A screen-shot of replicate MEL18 ChIP-seq profiles around ALX4 PTE from (B). Positions of corresponding MEL18 peak summits (marked with red and green dashed lines) deviate by 194 bp due to experimental variance. The median genomic position between paired peaks is assigned as the center of candidate ALX4 PTE. The half distance between the peaks estimates the accuracy of the candidate PTE location. (B) H3K27me3, SUZ12, RING2 and MEL18 ChIP-seq profiles over ALX4 locus, which harbors a candidate PTE (red rectangle, ChIP-qPCR amplicon indicated with black line) marked by obvious MEL18 peak. The heat-map underneath ChIP-seq profiles shows the number of CpG nucleotides within the 100 bp sliding window (ranging from dark blue = 0 to bright yellow = 15). (C) The outline of the transgenic assay. To increase the throughput of the assay, the cloning procedure was modified to include Gateway attL/attR recombination (yellow and blue arrows). Cells were transduced with pools of up to six different lentiviral constructs. As in earlier experiments, a small stretch of nucleotides (red circle) within the cloned fragment (green arch) was replaced to distinguish the transgenic and endogenous copies.