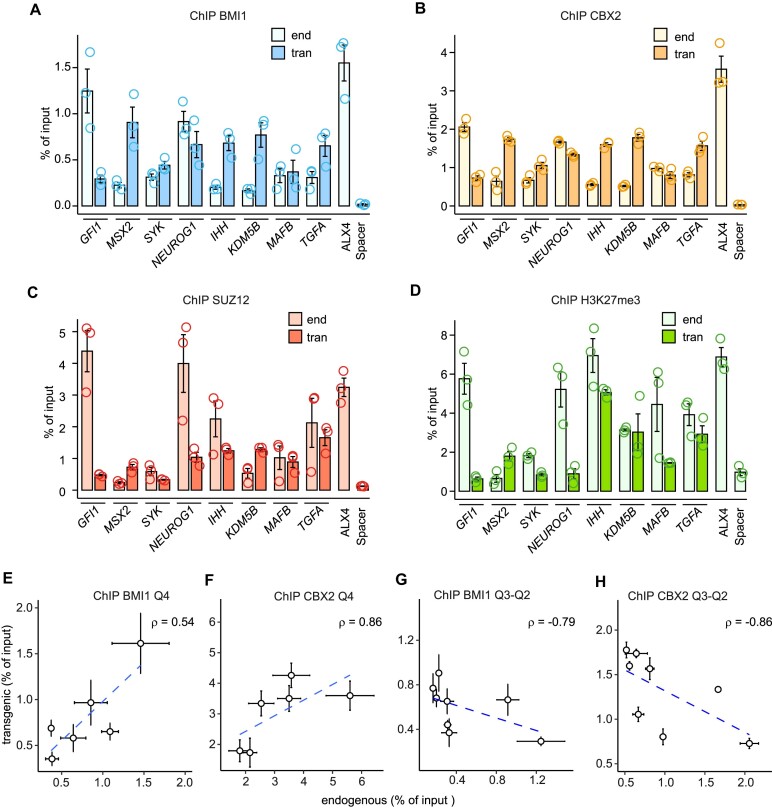
Figure 6.
PRC1 tethering by low-occupancy sites. ChIP-qPCR experiments indicate that DNA fragments underneath selected Q3 and Q2 MEL18 peaks always generate new binding sites for BMI1 (A) and CBX2 (B) when integrated elsewhere in the genome. Sometimes they also generate new binding sites for SUZ12 (C) and H3K27me3 (D). Immunoprecipitation of ALX4 and an intergenic region (Spacer) were used as positive and negative controls, respectively. Comparison of BMI1 (E, G) and CBX2 (F, H) ChIP-qPCR signals at transgenic and endogenous locations indicate that those, corresponding to the high-occupancy PTEs, correlate but those, corresponding to the low occupancy MEL18 peaks, do not. The data for high-occupancy PTEs is from Figure 3A, B. The whiskers show the standard error of the mean for three independent experiments.