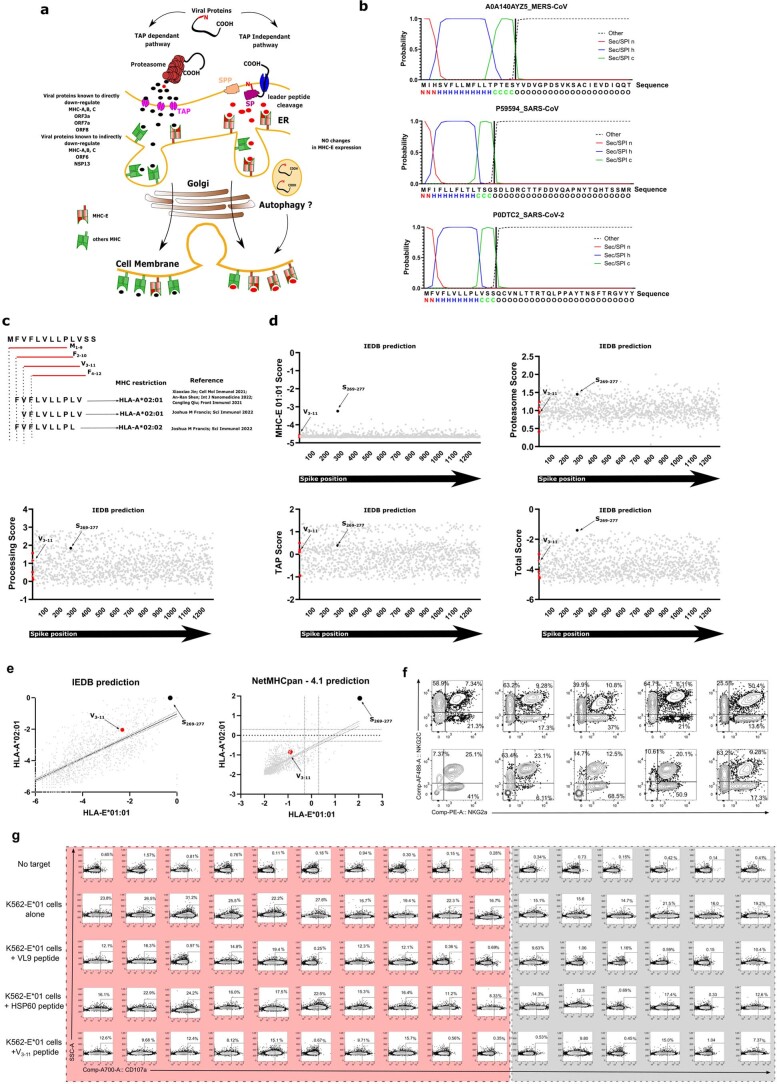
Extended Data Fig. 9. Identifying HLA-E Binding Potential in SARS-CoV-2 Signal Peptides.
(a) Literature-based scheme summarizing peptide processing via TAP-dependent and TAP-independent pathways. (b) In silico analysis predicting signal peptide presence in spike proteins and their cleavage sites, with SignalP 6.0 software suggesting cleavage by Signal peptidase (SP) and Signal peptide peptidase (SPP). Signal peptides comprise hydrophobic core (h region), polar C-terminal end (c region), and polar N-terminal region (n region). (c) Schematic displaying four overlapping nonamers in the 5’ region of SARS-CoV-2 signal peptide (P0DTC2), with V3-11 resembling the MHC-E binding motif. MHC restrictions and references provided. (d) In silico epitope predictions for HLA-E binding using the IEDB analysis resource, covering the entire spike sequence. Graphs exhibit HLA-E*01:01 binding scores, proteasomal cleavage, transport scores, and total scores combining HLA-E binding and processing. Red dots represent spike SP peptides from (c), and S269–277 corresponds to a previously described peptide. (e) Comparison of binding probabilities to HLA-E and HLA-A using NetMHCpan - 4.1, analyzing the entire SARS-CoV-2 spike protein. Multiple nonamers represented as black dots, with dot positions indicating predicted HLA-E binding capacity. Percentile ranks for HLA-E01:01 (y-axis) and HLA-E02:01 (x-axis) are provided. (f) Representative dot plot showcasing NKG2A+ and NKG2C+ human NK cell responses. (g) Dot plots displaying the percentage of CD107a+ NK cells isolated from HC and WTM at ≥ 221 days p.i. after co-culture with K562-E*01 cells loaded with peptides (VL9, HSP60, or V3-11) or no peptides. Light squares indicate wildtype infected convalescent monkeys, while light gray squares represent HC.