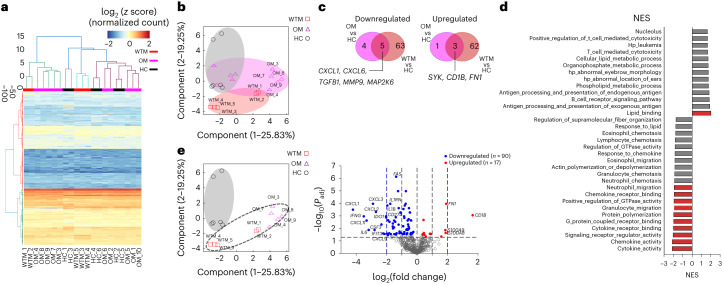
Fig. 3. SARS-CoV-2 replication in Mac in vitro alters Mac transcriptomic profile.
a, Heatmap for mRNA expression in BALF CD64+ Mac isolated from 5 HC, 5 randomly chosen WTM and 10 OM > 221 d p.i., cultured for 8 h and profiled with the nanoString nCounter System. Cluster distance values were determined with the Euclidean method. b, PCA of gene expression in BALF macrophages as in a. c, Venn diagram showing the genes commonly downregulated and upregulated in BALF MAC (as in Fig. 2a) of WTM and OM compared to HC counterpart. d, GSEA hallmark analysis showing enriched gene sets based on the differentially regulated genes from Fig. 2a. Red bars indicate significant enrichment at FDR < 25%, gray bars represent gene sets with FDR > 25% and a nominal P value < 5%. A positive NES value indicates enrichment in WTM and OM, and a negative NES indicates enrichment in HC. e, PCA and volcano plots showing upregulated and downregulated genes for the indicated comparison. In each volcano plot, the horizontal dotted line represents a Padj = 0.05 and the vertical dotted line represents a log(fold change) >2 or <−2. NES, normalized enrichment score.