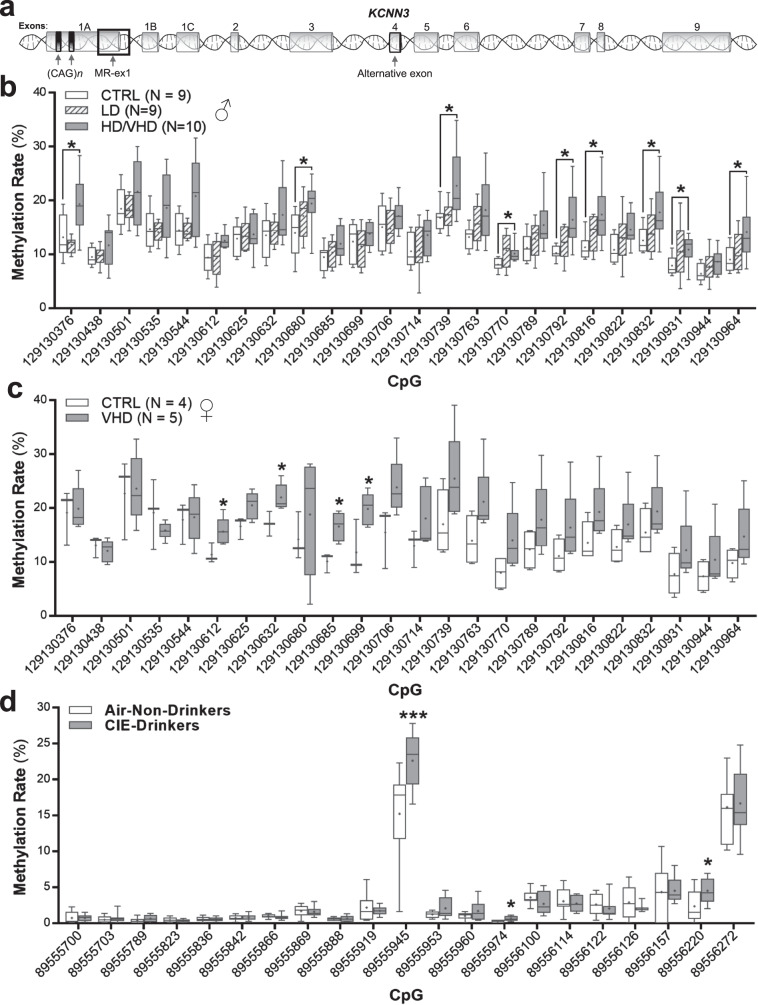
Fig. 3. KCNN3 methylation levels within MR-ex1-200 of ethanol-drinking monkeys and dependent mice.
The average DNAm rates (±SEM) of individual CpGs included in the methylation region under study are shown. a Exon organization of the KCNN3 locus showing the location of exons and the dual CAG trinucleotide repeat arrays and methylated region in exon 1 (MR-ex1). b In male rhesus macaques, the following CpGs showed elevated rates of DNAm in heavy/very heavy drinking macaques vs controls: CpG129130376: F(2, 11.846) = 7.9, *p = 0.04; CpG129130680: F(2, 15.955) = 3.661, *p = 0.036; CpG129130739: F(2, 15.468) = 3.817, *p = 0.04; CpG129130770: F(2, 13.57) = 5.047, *p = 0.041; CpG129130792: F(2, 13.945) = 7.047, *p = 0.01; CpG129130816: F(2, 15.63) = 5.836, *p = 0.015; CpG129130832: F(2, 15.473) = 3.95, *p = 0.033; CpG129130931: F(2, 14.393) = 4.016, *p = 0.038; CpG129130964: F(2, 15.708) = 3.985, *p = 0.031. c In female macaques, elevated rates of methylation were observed at the following CpGs in very heavy drinking macaques vs controls: CpG129130612: F(1, 7) = 6.122, *p = 0.048; CpG129130632: F(1, 7) = 7.64, *p = 0.033; CpG129130685: F(1, 7) = 13.370, *p = 0.011; CpG129130699: F(1, 7) = 7.799, *p = 0.031. d In mouse NAcC, the following CpGs showed elevated rates of DNAm between non-drinking mice and drinking-dependent mice: Independent t-tests: CpG89555945, t(16)= −2.946, ***p = 0.0009; CpG89555974, t(17) = −2.860, *p = 0.011; CpG89556220, t(17) = −2.206, *p = 0.042.