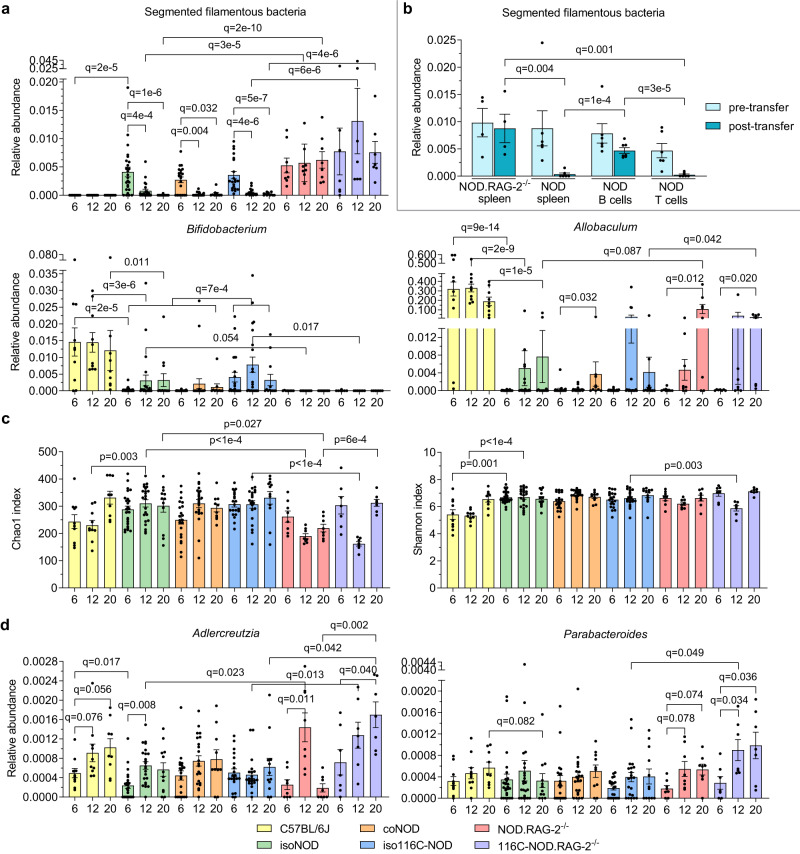
Fig. 8. Relative abundance and α-diversity of gut microbiota significant taxa from immunocompetent and immunodeficient mouse models.
16S rRNA gene analysis was performed in faecal samples at 6, 12, and 20 weeks of age of different mouse strains: control C57BL/6J (6 weeks: n = 10, 12 weeks: n = 10, 20 weeks: n = 9), isolated NOD or isoNOD (6 weeks: n = 25, 12 weeks: n = 23, 20 weeks: n = 12), cohoused NOD or coNOD (6 weeks: n = 21, 12 weeks: n = 22, 20 weeks: n = 10), isolated 116C-NOD or iso116C-NOD (6 weeks: n = 21, 12 weeks: n = 21, 20 weeks: n = 12), NOD.RAG-2−/− (6 weeks: n = 8, 12 weeks: n = 8, 20 weeks: n = 8), and 116C-NOD.RAG-2−/− (6 weeks: n = 7, 12 weeks: n = 7, 20 weeks: n = 7). a Relative abundance of significant taxa altered by the autoimmune diabetes lymphocyte milieu. b Relative abundance of segmented filamentous bacteria before (pre-transfer, 6 weeks) and after the transfer (post-transfer, 12 weeks) of total NOD spleen (n = 6), NOD B cells (n = 7), NOD T cells (n = 7), and control NOD.Rag2−/− spleen (n = 4). c α-diversity based on Chao1 and Shannon indexes. d Relative abundance of bacteria affected by 116C-NOD B cells. a, c, and d share the same legend. Data are expressed as mean ± SE. Relative abundance data were analysed using the MaAsLin2 statistical framework (mixed-effects linear regression model, two-sided test, adjustment for multiple comparisons), where p-values were corrected using the false discovery rate (FDR). Chao1 and Shannon indexes were analysed with Mann–Whitney’s test (two-tailed p-values).