Abstract
Phylogenetic analysis of 16S ribosomal DNA (rDNA) clones obtained by PCR from uncultured bacteria inhabiting a wide range of environments has increased our knowledge of bacterial diversity. One possible problem in the assessment of bacterial diversity based on sequence information is that PCR is exquisitely sensitive to contaminating 16S rDNA. This raises the possibility that some putative environmental rRNA sequences in fact correspond to contaminant sequences. To document potential contaminants, we cloned and sequenced PCR-amplified 16S rDNA fragments obtained at low levels in the absence of added template DNA. 16S rDNA sequences closely related to the genera Duganella (formerly Zoogloea), Acinetobacter, Stenotrophomonas, Escherichia, Leptothrix, and Herbaspirillum were identified in contaminant libraries and in clone libraries from diverse, generally low-biomass habitats. The rRNA sequences detected possibly are common contaminants in reagents used to prepare genomic DNA. Consequently, their detection in processed environmental samples may not reflect environmentally relevant organisms.
Knowledge of microbial diversity has increased dramatically in recent years, in part as a result of sequencing of rRNA genes from DNA obtained directly from uncultured microbiota, often by use of PCR and rRNA-specific primers (2, 21). This approach has been applied to assess the microbial diversity in a variety of environments, for instance, arctic tundra (34), marine deep subsurfaces (27), Yellowstone hot springs (14), peat bogs (26), and human infections (9, 12, 17). Although the approach has produced a diverse collection of sequences and expanded our view of microbial diversity, analysis of microbial 16S ribosomal DNA (rDNA) sequences has limitations in relating specific rDNA sequences to organisms in the environment under study and is fraught with potential artifacts.
One potential artifact in the application of PCR to community analysis is the possible introduction of contaminating rDNA during experimental procedures, particularly in steps preceding PCR. In the course of compiling environmental 16S rDNA sequences, we have noted some highly similar (>99%) sequences that are obtained from many physically and chemically distinct environments. The organisms represented by these sequences may indeed be prevalent in such diverse environments. However, the difficulty in preparing genomic DNA absolutely free from contaminating DNA, coupled with the exquisite sensitivity of PCR to amplify trace target DNA, make contamination a serious issue, particularly with low-biomass samples (16, 18, 28, 29). We report here a survey of 16S rDNA sequences that were obtained from negative extraction controls, that is, DNA extraction and purification performed without added sample, and the correspondence of these sequences to some recovered from diverse environmental settings.
We surveyed the 16S rDNA sequences from 96 clones derived from a PCR product resulting from a control extraction that did not contain an environmental sample. Less-extensive analyses have been conducted with independently processed negative controls. The extraction was carried out in the same manner as with authentic samples containing genomic DNA, by using lysozyme, proteinase K, sodium dodecyl sulfate, bead beating, and phenol treatment as described elsewhere (4). Details were as follows: buffer A consisted of 200 mM Tris HCl (pH 8.0), 200 mM NaCl, and 20 mM EDTA, bead beating (0.5 g of acid-washed beads) was carried out for 2 min at low speed and 0.5 min at high speed, and nucleic acids were precipitated from 300 mM NaCl and 3 volumes of 100% ethanol. All solutions were prepared with autoclaved, filtered (0.2-μm-pore-size filter, Sterile Acrodisc; Gelman Sciences) ddH2O prior to use. Although some small bacteria potentially could pass through the 0.2-μm filter pores, a negative control in the absence of any template, that is, without the negative extraction control material, resulted in no PCR product after 40 cycles. All DNA extraction procedures and manipulations were performed in a laminar flow hood to minimize aerial contamination. The sample was dissolved in 200 μl of water, and 100-μl PCRs were carried out with 1 μl of template (40 cycles of touchdown PCR, consisting of 20 cycles of 1 min at 94°C, 40 s starting at 67°C and decreasing by 1°C/cycle, and 1 min at 72°C, and 20 cycles of 1 min at 94°C, 1 min at 50°C, and 1.5 min plus 1 s/cycle at 72°C). Five units of Ampli Taq Gold (Perkin-Elmer) was added per 100 μl of reaction mixture. Bacterium-specific primers were used for the amplification (27F, AGAGTTTGATCCTGGCTCAG, and 805R, GACTACCAGGGTATCTAATCC). These primers match most sequences in the domain Bacteria in the primer target region. Three 100-μl reaction products were precipitated with ethanol, and the entire sample was resolved by agarose gel electrophoresis. Even with these precautions, rDNA-sized PCR products were obtained after extended rounds of thermal cycling. The rDNA-sized PCR products were eluted from the gel and cloned, and 96 clones were analyzed by restriction fragment length polymorphisms (RFLP) by using the enzymes MspI and HinP1I as detailed by Hugenholtz et al. (14). Twenty different RFLP types were identified and sequenced. The 16S rDNA sequences (460 to 780 nucleotides [nt]) were compared to known sequences by using the gapped BLAST search algorithm (1, 5) and were aligned to close relatives by using the GDE alignment editor (19). 16S rDNA sequences were placed into a phylogenetic tree containing more than 7,000 bacterial rDNA sequences by using the ARB software package (30).
Analysis of clones from the negative extraction control revealed a diverse collection of contaminant sequences. We have seen similar sequences in other analyses of contaminant rDNAs. 16S rDNA sequences essentially identical to the negative-control sequences also have been encountered by a number of laboratories in clone libraries from a broad diversity of environments, as summarized in Table 1. Additional sequences from the negative control, not reported in environmental analyses, are related to the rDNAs of the following organisms: Micrococcus luteus (98% identity to MT2), Pseudomonas aeruginosa (99% identity to MT5), an Afipia sp. (97% identity to MT8), Variovorax paradoxus (97% identity to MT11), Gemella haemolysans (100% identity to MT1), Shigella boydii (99% identity to MT19), and Phyllobacterium myrsinacearum (99% identity to MT17). A few sequences were <95% identical to those of known organisms. A compilation of sequences obtained from negative-control libraries is available from our web site at http://crab2.berkeley.edu/pacelab/177.htm. This site will be updated as additional sequences are determined from negative-control libraries. Submissions are welcomed.
TABLE 1.
Distribution of contaminant clones with representatives from environmental studies
| Clone(s) from negative control | Environment and clone >98% identical to negative-control clone (reference) | GenBank no. | Most closely related organism | % Iden- titya |
|---|---|---|---|---|
| MT3 | Bentonite/sand subsurface, Canada, clone K13 (22) | X91528 | Stenotrophomonas maltophilia | 96.7 |
| Acid mine drainage, clone TRA2-7 (11) | AF047649 | |||
| MT6 | Bentonite/sand subsurface, Canada, clone K9 (22) | X91524 | Acinetobacter sp. DSM590 | 99.7 |
| Deep marine sediment, clone JAP752 (27) | U09828 | |||
| Groundwater/subsurface at Äspö hard rock lab, Sweden, clone A24otpmn (23) | X91447 | |||
| Deep granitic groundwater, Stripa research mine, Sweden, clone Group III (10) | L20812 | |||
| MT9 | Bentonite/sand subsurface, Canada, clone K16 (22) | X91531 | Escherichia coli | 98.0 |
| Prostatitis, clone 5725 (17) | NAb | |||
| MT14, CMT35P | Groundwater/subsurface, Gabon, Africa, clone G4 (24) | X91175 | Leptothrix cholodnii | 96.5 |
| Infected guinea pig lung, clone GPMT16 (31) | AF058793 | |||
| Acid mine drainage, clone TRB32 (11) | AF047647 | |||
| MT18, CTHB-18 | Bentonite/sand subsurface, Canada, clone K11 (22) | X91526 | Duganella zoogloeoides (formerly Zoogloea ramigera) | 99.8 |
| Deep marine sediment, clone JAP405 (27) | U09778 | |||
| Acid mine drainage, clone AMDke9.1 (11) | NA | |||
| Antarctic, clone BP-S155 (11) | NA | |||
| Basalt sand deep subsurface, clone BS43 (11) | NA | |||
| Yellowstone hot spring, clone OPS122B (14) | AF026984 | |||
| Siberian tundra soil, clone S-41 (34) | AF016752 | |||
| Dentoalveolar abscess (pus sample), clone PUS 10.40 (9) | U34035 | |||
| Contaminated aquifer, clone WFeA1-06 (8) | AF050528 | |||
| Groundwater/subsurface, Gabon, Africa, clones G22 and G41 (24) | X91274 | |||
| Hawaiian soil, clone HRS-17 (20) | AF016530 | |||
| Humic-reducing environment, molecular isolate from a PCR product (6) | AF019941 | |||
| Guinea pig lung, clone MTseq15 (31) | AF058388 | |||
| Mycobacterium tuberculosis-infected guinea pig, lung, clone GPMT3 (31) | AF058387 | |||
| MT22 | Low-pH peat bog, clones TM221 and TM252 (26) | X97095 | Herbaspirillum seropedicae | 98.6 |
| Carolina bay sediment, clones RB-06 and RB-37 (33) | U62830 |
Between the negative-control clone and the related organism.
NA, not available.
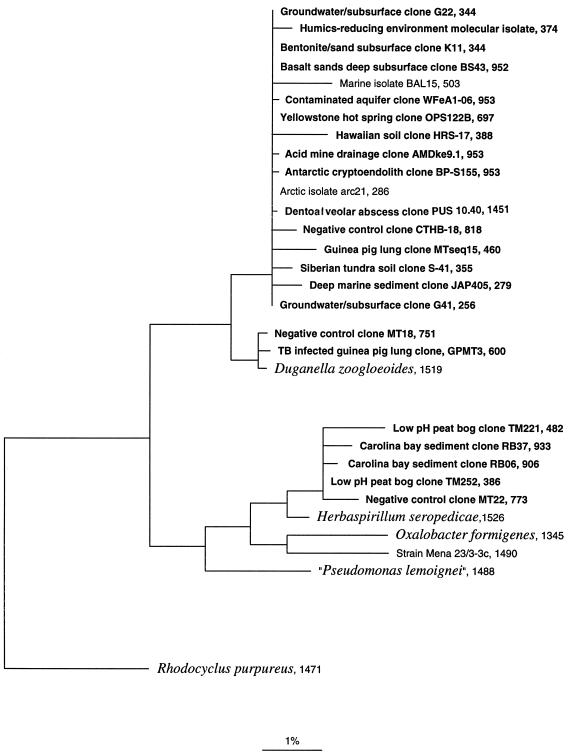
Several contaminant clones and numerous environmental clones from diverse environments are closely related to rDNAs of the genus Duganella (formerly Zoogloea). Such organisms (β-proteobacteria) commonly contaminate water sources and are routinely isolated from wastewater environments (13) and drinking-water biofilms (15). Their occurrence in materials used for laboratory experiments is, therefore, not surprising. Figure 1 shows the relationships of some of the contaminant sequences and environmental groups of sequences to those of Duganella zoogloeoides and Herbaspirillum seropedicae. The Duganella relatedness group, seen as two clades with ca. 98.5% identity in rRNA sequences, consists mostly of environmental rDNA sequences, all nearly identical, from various published studies (6, 8, 9, 20, 22–24, 27, 34). In addition to the Duganella cluster of sequences, a number of other prevalent contaminant sequences, listed in Table 1, correspond closely to sequences reported from diverse environments. Leptothrix spp., for instance, like Duganella spp., are found in slowly flowing fresh water and in polluted water and activated sludge. It is remarkable that essentially identical rRNA sequences are obtained from such different environments as deep subsurface groundwaters, a marine sediment, a Yellowstone hot spring, a guinea pig lung, and a dentoalveolar abscess (pus around the teeth). All of the extractions of environmental samples that resulted in clones equivalent to the contaminant rDNAs are likely to have contained only very low levels of biomass. Extraction of DNA from low-biomass samples is particularly sensitive to potentially contaminating DNA during processing because the contaminating DNA is minimally diluted by sample DNA. The exact sources of contamination were not determined; however, the Taq polymerase and amplification buffer are not detectably contaminated, since reactions without added template or negative extraction control material did not produce amplified products. Therefore, the most likely sources of contamination are salts and buffers, lysozyme, proteinase K, and/or the zirconia/silica beads.
FIG. 1.
Evolutionary-distance dendrogram showing the relative positions of environmental 16S rDNA clone and strain sequences to the genera Duganella and Herbaspirillum. Bar indicates nucleotide substitution rate. The sequence length in nucleotides follows the clone or strain designation. References for clones are given in Table 1. Clones are boldfaced; the cultivated strains marine isolate BAL15 (25), arctic isolate arc21 (3), and strain Mena 23/3-3c (32) are shown in lightface. Sequences for described bacterial species (italics) were obtained from GenBank (5). The rDNA sequence from Rhodocyclus purpureus was used as an outgroup. The resolution of the tree shown here is limited by the quality of the sequence data and the short lengths (fewer than 500 nt) of many sequences. Short sequences (<500 nt) were inserted into the tree by using the parsimony insertion tool of the ARB software program (30). The contaminant clone CTHB-18 was identified in a separate negative-control extraction independently from that for the MT clone library.
In this study, we report that many small-subunit rRNA sequences obtained from no-sample control clone libraries are closely related to sequences recovered in other studies from diverse environmental samples. This correspondence of contaminant and environmental sequences may indicate that some of the environmental sequences are derived as experimental contaminants and have no relevance to the environmental communities. Since any source of contaminant sequences is likely to be idiosyncratic, dependent on the operator, source of reagents, water, etc., researchers examining biodiversity using environmental cloning techniques must be aware of this issue and make every effort to minimize, detect, and analyze potential contamination. Perhaps the most important message from results presented here is that definitive proof for the occurrence of an organism indicated by a cloned rRNA sequence requires explicit identification of that organism in situ. Currently, this is most readily achieved by using 16S rRNA-based fluorescence hybridization techniques (2, 7).
Nucleotide sequence accession numbers.
Sequences for the following negative-control clones (with the accession numbers given in parentheses) were deposited in the GenBank database: MT3 (AF058381), MT6 (AF058382), MT9 (AF058375), MT11 (AF058383), MT14 (AF058384), MT18 (AF058385), MT22 (AF058386), CMT35P (AF061574), and CTHB-18, (AF067655). MT2, MT5, and MT8 have accession no. AF058372 to AF058374, respectively. MT12, MT17, and MT19 have accession no. AF058377 to AF058379, respectively.
Acknowledgments
We thank Phil Hugenholtz for comments on the manuscript and Chris Pitulle for stimulating discussions.
This work was supported by grants from the NIH and the U.S. Department of Energy.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahr M, Hobbie J E, Sogin M L. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- 4.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coates J D, Ellis D J, Blunt-Harris E L, Gaw C V, Roden E E, Lovley D R. Recovery of humic-reducing bacteria from a diversity of environments. Appl Environ Microbiol. 1998;64:1504–1509. doi: 10.1128/aem.64.4.1504-1509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 8.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. Microbial diversity in a hydrocarbon and chlorinated solvent-contaminated aquifer undergoing intrinsic bioremediation. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 9.Dymock D, Weightman A J, Scully C, Wade W G. Molecular analysis of microflora associated with dentoalveolar abscesses. J Clin Microbiol. 1996;34:537–542. doi: 10.1128/jcm.34.3.537-542.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekendahl S, Arlinger J, Ståhl F, Pedersen K. Characterization of attached bacterial populations in deep granitic groundwater from the Stripa research mine by 16S rRNA gene sequencing and scanning electron microscopy. Microbiology. 1994;140:1575–1583. doi: 10.1099/13500872-140-7-1575. [DOI] [PubMed] [Google Scholar]
- 11.Goebel, B. M., and N. R. Pace. 1998. Unpublished data.
- 12.Goldenberger D, Künzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiraishi A, Shin Y K, Sugiyama J. Proposal to reclassify Zoogloea ramigera IAM 12670 (P. R. Dugan 115) as Duganella zoogloeoides gen. nov., sp. nov. Int J Syst Bacteriol. 1997;47:1249–1252. doi: 10.1099/00207713-47-4-1249. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalmbach S, Manz W, Szewzyk U. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl Environ Microbiol. 1997;63:4164–4170. doi: 10.1128/aem.63.11.4164-4170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitchin P A, Szotyori Z, Fromholc C, Almond N. Avoidance of PCR false positives. Nature. 1990;344:201. doi: 10.1038/344201a0. [DOI] [PubMed] [Google Scholar]
- 17.Krieger J N, Riley D E, Roberts M C, Berger R E. Prokaryotic DNA sequences in patients with chronic idiopathic prostatitis. J Clin Microbiol. 1996;34:3120–3128. doi: 10.1128/jcm.34.12.3120-3128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 19.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nüsslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen K. Investigations of subterranean bacteria in deep crystalline bedrock and their importance for the disposal of nuclear waste. Can J Microbiol. 1996;42:382–391. [Google Scholar]
- 23.Pedersen K, Arlinger J, Ekendahl S, Hallbeck L. 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Äspö hard rock laboratory, Sweden. FEMS Microbiol Ecol. 1996;19:249–262. [Google Scholar]
- 24.Pedersen K, Arlinger J, Hallbeck L, Pettersson C. Diversity and distribution of subterranean bacteria in groundwater at Oklo in Gabon, Africa, as determined by 16S rRNA gene sequencing. Mol Ecol. 1996;5:427–436. doi: 10.1111/j.1365-294x.1996.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 25.Pinhassi J, Zweifel U L, Hagström Å. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 27.Rochelle P A, Cragg B A, Fry J C, Parkes R J, Weightman A J. Effect of sample handling on estimation of bacterial diversity in marine sediments by 16S rRNA gene sequence analysis. FEMS Microbiol Ecol. 1994;15:215–226. [Google Scholar]
- 28.Sarkar G, Sommer S S. Shedding light on PCR contamination. Nature. 1990;343:27. doi: 10.1038/343027a0. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt T M, Pace B, Pace N R. Detection of DNA contamination in Taq polymerase. BioTechniques. 1991;11:176–177. [PubMed] [Google Scholar]
- 30.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Struckmann, B. Nonhoff, M. Lenke, A. Vilbig, T. Ludwig, A. Bode, K. H. Schleifer, and W. Ludwig. ARB: a software environment for sequence data. Submitted for publication.
- 31.Tanner, M. A., and N. R. Pace. 1998. Unpublished data.
- 32.Tinschert A, Kiener A, Heinzmann K, Tschech A. Isolation of new 6-methylnicotinic-acid-degrading bacteria, one of which catalyses the regioselective hydroxylation of nicotinic acid at position C2. Arch Microbiol. 1997;168:355–361. doi: 10.1007/s002030050509. [DOI] [PubMed] [Google Scholar]
- 33.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]