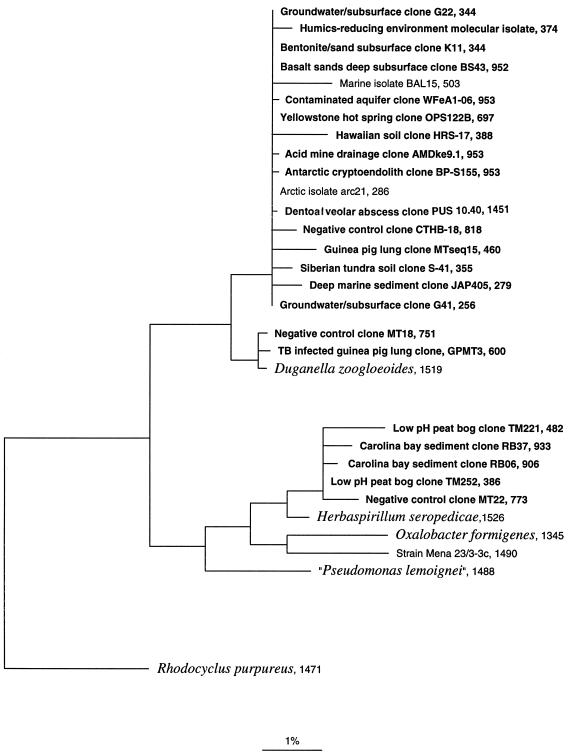
FIG. 1.
Evolutionary-distance dendrogram showing the relative positions of environmental 16S rDNA clone and strain sequences to the genera Duganella and Herbaspirillum. Bar indicates nucleotide substitution rate. The sequence length in nucleotides follows the clone or strain designation. References for clones are given in Table 1. Clones are boldfaced; the cultivated strains marine isolate BAL15 (25), arctic isolate arc21 (3), and strain Mena 23/3-3c (32) are shown in lightface. Sequences for described bacterial species (italics) were obtained from GenBank (5). The rDNA sequence from Rhodocyclus purpureus was used as an outgroup. The resolution of the tree shown here is limited by the quality of the sequence data and the short lengths (fewer than 500 nt) of many sequences. Short sequences (<500 nt) were inserted into the tree by using the parsimony insertion tool of the ARB software program (30). The contaminant clone CTHB-18 was identified in a separate negative-control extraction independently from that for the MT clone library.