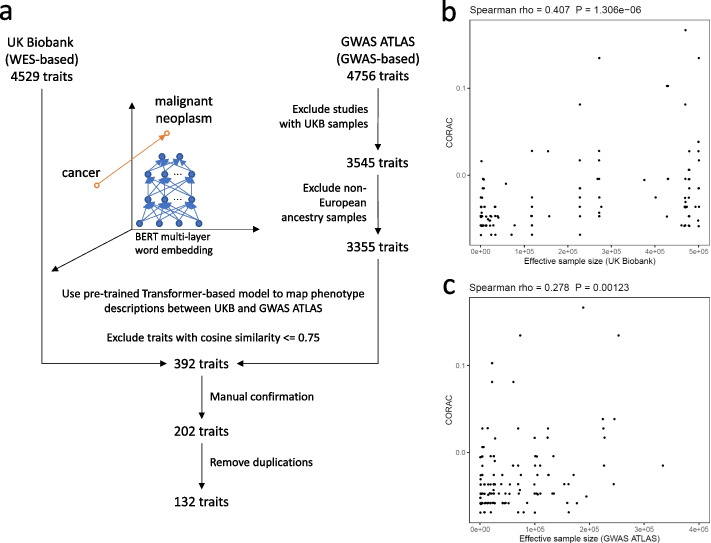
Fig. 6.
Independent data sources for the common and rare variant-based signals verified the correlation between CORAC and effective sample size. Instead of the UK Biobank as the data source for common variant-based signals, we leveraged the data from GWAS ATLAS to investigate the impact of the use of a shared dataset from which the common and rare variant signals were derived. Here, GWAS data that included any samples from the UK Biobank were excluded from the data source for the common variant-based signals. A pretrained Sentence-BERT word embedding model was implemented. The Transformer-based network searches for semantic similarity, enabling the mapping of phenotype descriptions in the UK Biobank to those in the GWAS ATLAS. Cosine similarity analysis (for semantic textual similarity), manual confirmation, and removal of duplications were then performed (a; Methods). The scatter plot shows the correlation between CORAC and effective sample size derived from the shared UK Biobank dataset (b) and from the independent data sources (c)