Mahone et al. use genetic, biochemical, and single-molecule imaging approaches to define a signaling pathway that coordinates chromosome segregation with progression of constriction during cell division in the bacterium Caulobacter crescentus. This pathway ensures genome integrity during the division process.
Abstract
To divide, bacteria must synthesize their peptidoglycan (PG) cell wall, a protective meshwork that maintains cell shape. FtsZ, a tubulin homolog, dynamically assembles into a midcell band, recruiting division proteins, including the PG synthases FtsW and FtsI. FtsWI are activated to synthesize PG and drive constriction at the appropriate time and place. However, their activation pathway remains unresolved. In Caulobacter crescentus, FtsWI activity requires FzlA, an essential FtsZ-binding protein. Through time-lapse imaging and single-molecule tracking of Caulobacter FtsW and FzlA, we demonstrate that FzlA is a limiting constriction activation factor that signals to promote conversion of inactive FtsW to an active, slow-moving state. We find that FzlA interacts with the DNA translocase FtsK and place FtsK genetically in a pathway with FzlA and FtsWI. Misregulation of the FzlA-FtsK-FtsWI pathway leads to heightened DNA damage and cell death. We propose that FzlA integrates the FtsZ ring, chromosome segregation, and PG synthesis to ensure robust and timely constriction during Caulobacter division.
Introduction
Bacterial cell division is a robust process that requires tight regulation of multiple events to ensure survival. These events include marking the division site, recruiting division proteins, segregating the chromosome, remodeling and synthesizing the cell wall, and separating the daughter cells (Dewachter et al., 2018; Mahone and Goley, 2020). The first step in division is the assembly of FtsZ, an essential and conserved tubulin homolog, into a cytokinetic “Z-ring” at the incipient division site (Mahone and Goley, 2020; Barrows and Goley, 2021). Once the Z-ring is established, dozens of proteins (the divisome) are directly or indirectly recruited by FtsZ to the division site (Mahone and Goley, 2020; McQuillen and Xiao, 2020). FtsZ polymers within the Z-ring are highly dynamic, driven by FtsZ’s GTPase activity, and exhibit treadmilling motion (Bisson-Filho et al., 2017; Yang et al., 2017).
After divisome assembly, cells constrict inward via envelope remodeling. In Gram-negative bacteria, the cell envelope comprises an inner membrane, cell wall, and outer membrane. The peptidoglycan (PG) cell wall is a meshwork that protects against turgor pressure and dictates bacterial morphology (Daitch and Goley, 2020). New PG is synthesized by glycosyltransferases that polymerize lipid II, a lipid-linked disaccharide made of N-acetylmuramic acid and N-acetylglucosamine with a pentapeptide side chain. The peptide stems are crosslinked by transpeptidases to create the PG mesh (Daitch and Goley, 2020). During division, the PG synthases FtsW and FtsI are the primary glycosyltransferase and transpeptidase, respectively (Daitch and Goley, 2020; Mahone and Goley, 2020). These enzymes work together as a cognate pair (FtsWI) to synthesize cytokinetic PG that provides the constrictive force (Coltharp and Xiao, 2017).
Single-molecule tracking (SMT) studies show that the PG synthases move dynamically about the division site. In bacteria where dynamics have been characterized (Escherichia coli, Bacillus subtilis, Staphylococcus aureus, and Streptococcus pneumoniae), the dynamics of FtsZ and the PG synthases are associated, but range from FtsWI requiring FtsZ treadmilling for movement to requiring FtsZ for placement at midcell, but moving independently (Yang et al., 2017, 2021; Bisson-Filho et al., 2017; Perez et al., 2019; Yang and Liu, 2022; Schäper et al., 2023, Preprint; Whitley et al., 2023, Preprint). In E. coli, the moving PG synthases can be differentiated into fast- and slow-moving populations. Fast-moving PG synthases depend on FtsZ for movement and are inactive for PG synthesis. Slow-moving molecules depend on PG synthesis for locomotion and are enzymatically active (Yang et al., 2017, 2021; McCausland et al., 2021; Lyu et al., 2022).
In most model bacteria, divisome components upstream of FtsWI activation have been identified, but their precise functions and mechanisms of signaling remain unclear. In E. coli, FtsN is proposed to trigger constriction initiation and is last to localize to the division plane (Weiss, 2015; Mahone and Goley, 2020; Lyu et al., 2022). Hyperactivating mutations in ftsB and ftsL, which encode subunits of the FtsQLB complex, result in shorter cells (Tsang and Bernhardt, 2015; Liu et al., 2015). These mutants, when combined with other genetic perturbations, bypass requirements for otherwise essential divisome components, suggesting they are activators of constriction (Park et al., 2020; Li et al., 2022; Du et al., 2016). Consistent with this idea, purified FtsQLB from Pseudomonas aeruginosa activates FtsWI in vitro (Marmont and Bernhardt, 2020), and structural and bioinformatic approaches suggest a model wherein FtsQLB supports an active conformation of FtsWI (Käshammer et al., 2023; Britton et al., 2023). While the final stages of activation are becoming clear, how FtsZ-proximal partners signal to promote FtsWI activation remains elusive.
In Caulobacter crescentus (hereafter Caulobacter), an α-proteobacterium in which divisome assembly has been well defined, FtsW is the last divisome component to arrive at the division plane prior to constriction (Goley et al., 2011). However, localization of FtsW is not sufficient for constriction. Recently, we implicated FzlA, an essential division protein conserved across α-proteobacteria, as a regulator of constriction (Lariviere et al., 2018, 2019; Goley et al., 2010). FzlA binds FtsZ and colocalizes with FtsZ in stalked and predivisional cells (Goley et al., 2010). Previous work identified residues on FzlA required for two essential activities: binding to FtsZ and an unknown activity in the C-terminal tail (Lariviere et al., 2018). When FzlA is depleted or either essential activity is disrupted, cells do not constrict, despite the remainder of the divisome localizing to rings (Lariviere et al., 2018; Goley et al., 2010).
A hyperactive mutant of FtsW (FtsWA246T) was identified in a screen for mutations that suppress the toxicity of overexpression of the cell division inhibitors sidA or didA in Caulobacter (Modell et al., 2014). Replacing wild-type (WT) ftsW with ftsWA246T renders fzlA non-essential, albeit with a slower constriction rate when compared with a strain-producing FzlA (Lariviere et al., 2019). This mutant is analogous to the A234T mutation in E. coli RodA, a homolog of FtsW involved in maintaining rod shape. The PG polymerase activity of E. coli RodAA234T was increased compared with WT RodA in vivo and in vitro, supporting the assertion that this is a hyperactivating mutation (Rohs et al., 2018). An ftsWI triple mutant in Caulobacter (ftsWF145L/A246TftsII45V, termed ftsW**I*), consisting of three mutations that each suppress overexpression of sidA or didA, constricts faster than the ftsWA246T mutant and renders fzlA non-essential with minimal effects on cell length (Modell et al., 2014; Lambert et al., 2018; Lariviere et al., 2019). These genetic data led us to propose that FzlA signals to activate FtsWI as an FtsZ-proximal step in the FtsZ-FtsWI activation pathway.
While we have established that FzlA is required for FtsWI activation in Caulobacter, it is unlikely that FzlA directly regulates FtsWI activity. The mechanism by which FzlA signals for FtsWI activation and the functional importance of regulation by FzlA is unknown. Here, we employed time-lapse imaging, genetics, biochemistry, and SMT to dissect the relationship between FzlA and FtsWI activity. We discovered that FzlA signals to promote conversion of inactive FtsW molecules to an active, slow-moving state. Overproducing FzlA leads to hyperconstriction by promoting activation of FtsWI, and this hyperactivation is toxic, particularly in the ftsW**I* background. The toxicity of FzlA overproduction is in part due to enhanced DNA damage via misregulation of a novel interaction between FzlA and FtsK, a division protein that segregates chromosomal termini during division (Wang and Lutkenhaus, 1998; Wang et al., 2006). These results unify divisome assembly, chromosome segregation, and activation of cytokinetic PG synthases into a single pathway, which requires proper regulation to ensure envelope and chromosomal integrity.
Results
Overproduction of FzlA accelerates constriction
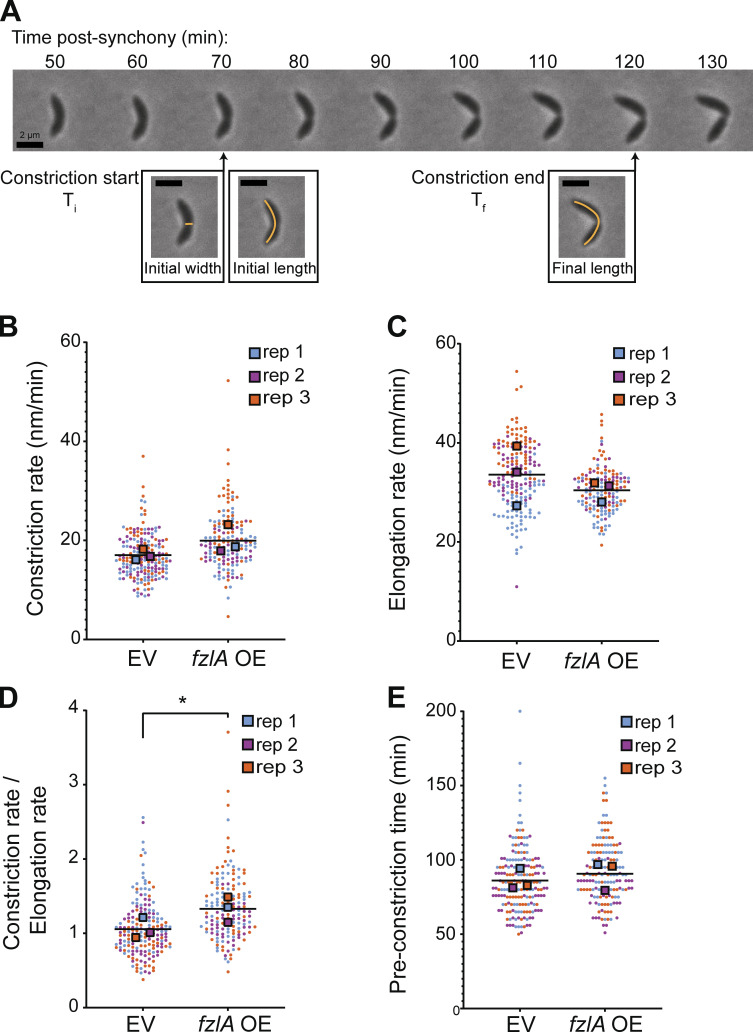
To understand how FzlA regulates FtsWI, we sought to characterize the effects of FzlA overproduction on division and viability. We hypothesized that FzlA may be a limiting factor in activation of FtsWI. If so, cells with excess FzlA should be hyperactive for constriction and, therefore, constrict faster than control cells (Lambert et al., 2018; Lariviere et al., 2019). To test this hypothesis, we performed time-lapse phase-contrast microscopy on synchronized cells overexpressing fzlA (EG3637) or bearing an empty vector (EV) control (EG1644; Fig. 1 A). We induced fzlA overexpression for 1 h prior to and throughout the time-lapse and isolated newborn swarmer (G1) cells by density centrifugation prior to imaging. We determined that fzlA-overexpressing cells increase FzlA levels to roughly 20-fold higher than WT (Fig. S1, A and B). For each cell, we calculated the rates of constriction and elongation, and the time from birth to initiation of constriction (preconstriction time). When FzlA was overproduced, cells constricted significantly faster on average (20.0 ± 0.5 nm/min, mean ± SEM, n = 154) than the EV control (16.8 ± 0.3 nm/min, n = 182) using conventional statistical analysis (Fig. 1 B and Video 1, P < 0.0001). To ensure that the measured differences were not due to sample size effects, we generated Superplots, treating the averages of independent biological replicates (rather than individual cell values) as data points for each condition (Lord et al., 2020). With Superplots analysis, the constriction rate was still significantly faster when fzlA was overexpressed (19.8 ± 1.6 nm/min) compared with an EV (16.9 ± 0.6 nm/min) using an α-cutoff of 0.1, but not when using an α-cutoff of 0.05 (Fig. 1 B, P = 0.1).
Figure 1.
Overproduction of FzlA hyperactivates constriction. (A) Representative phase-contrast time-lapse microscopy of a WT constricting Caulobacter cell. The timepoints of constriction initiation and completion are denoted. Scale bars, 2 µm. The orange lines represent measures of length or width for each cell. Ti: Time of initiation. Tf: Final timepoint of constriction. (B–E) Superplots comparing the (B) constriction rate (P = 0.1058, one-tail t test), (C) elongation rate (P = 0.2197, one-tail t test), (D) ratio of constriction to elongation rate (P = 0.0493, one-tailed t test), or (E) time to constriction initiation for strains harboring either an EV (EG1644) control or fzlA overexpression (OE; EG3637) construct (P = 0.6, Kolmogorov-Smirnov test). The strains were induced with 0.3% xylose for 1 h prior to and throughout the time-lapse experiment. Each circle is the value of a single cell, while the large squares are the average of a biological replicate. The bar is the average of the three means and statistics were performed using the means of the three biological replicates. EG1644—replicate 1, 67 cells; replicate 2, 63 cells; replicate 3, 54 cells. EG3637—replicate 1, 60 cells; replicate 2, 40 cells; replicate 3, 55 cells. *P < 0.05.
Figure S1.
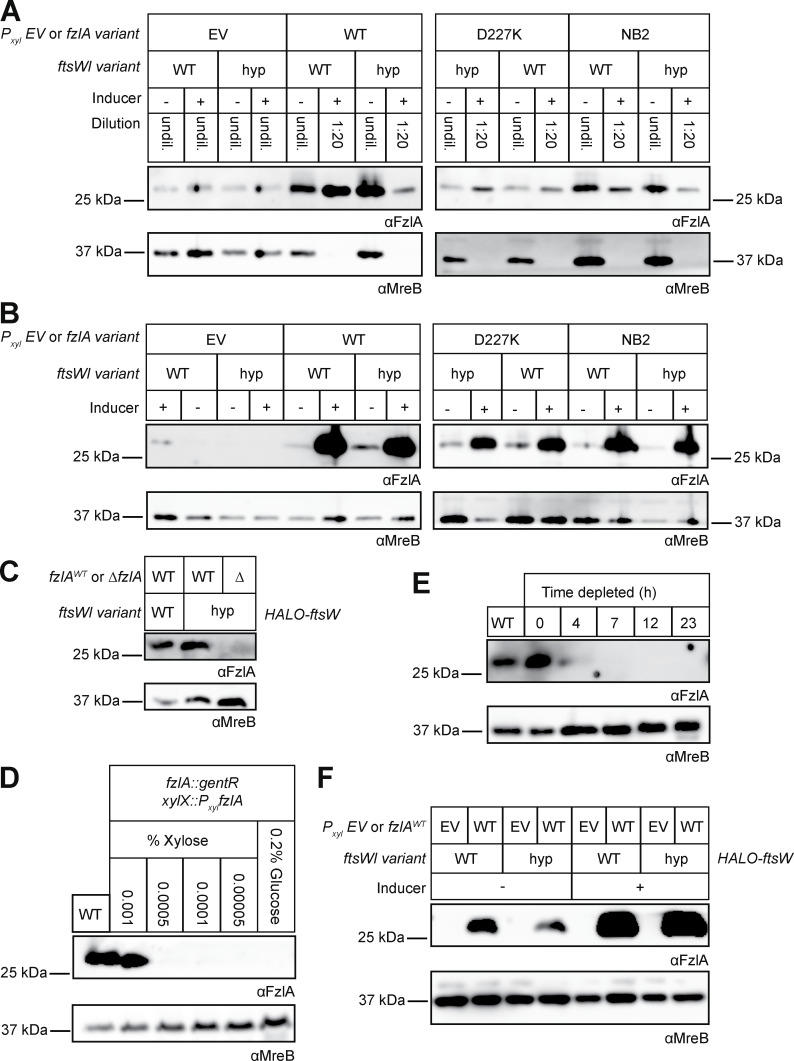
Western blots to confirm FzlA levels during strain characterization. (A–F) Western blot analysis using primary antibody recognizing either FzlA (αFzlA) or the loading control MreB (αMreB). (A and B) Overexpression of fzlA variants—WT FzlA (WT), a C-terminal tail mutant (D227K), or a mutant unable to bind FtsZ (NB2)—in WT and ftsW**I* (hyp) backgrounds compared with (EV) controls. (A) Strains with high FzlA were diluted 1:20 before loading as indicated to enable detection of all samples at a single exposure time. (B) Same as A but samples were all undiluted. (C) FzlA is not detected in the ∆fzlA; ftsW**I* strain. (D) fzlA induction in the halo-ftsW background (EG3523) as a function of xylose concentration to match FzlA levels in the WT halo-ftsW (EG3052) background. We selected 0.001% xylose as the closest approximation of WT FzlA levels. (E) FzlA depletion in the halo-ftsW background (EG3523). (F) FzlA overproduction in the halo-ftsW (EG3519) and halo-ftsW**; ftsI* (EG3525) backgrounds, as well as the respective EV controls (WT: EG3537. ftsW**I*: EG3538). Source data are available for this figure: SourceData FS1.
Video 1.
A WT division that completes successfully. Representative time-lapse phase-contrast microscopy of a constricting WT cell with an EV control. Each frame is 5 min apart and the total time-lapse is 4 h.
In Caulobacter, the elongation rate is inversely proportional to the constriction rate: as cells constrict faster, they elongate slower, and vice versa (Lambert et al., 2018; Lariviere et al., 2019). Superplots analysis of the elongation rates showed no significant difference between fzlA-overexpressing (15.2 ± 0.6 nm/min) and EV control cells (16.8 ± 1.7 nm/min; Fig. 1 C). In contrast, the ratio of constriction rate to elongation rate by Superplots analysis was significantly shifted in favor of constriction in strains overproducing FzlA (1.33 ± 0.10) when compared with control cells (1.06 ± 0.08), signifying a shift from elongation to constriction, similar to observations made for hyperactive strains (Fig. 1 D, P = 0.049; Lariviere et al., 2019). These results demonstrate that fzlA overexpression potentiates constriction. Notably, there was no significant difference in time from birth to constriction initiation between fzlA overexpressing (92 ± 2 min, n = 154) and EV control cells (86 ± 2 min, n = 182), even by conventional statistics (Fig. 1 E, P = 0.071). These data indicate that excess FzlA does not prematurely initiate constriction, but rather causes FtsWI to build PG inward at an increased rate.
Caulobacter FtsW moves in two populations: Slow, active molecules and fast, inactive molecules
We hypothesize that fzlA-overexpressing cells constrict faster because FtsWI are synthesizing PG more frequently or at an increased rate. We sought to directly test this assertion and reasoned that if Caulobacter FtsW dynamics are similar to E. coli FtsW dynamics (Yang et al., 2021), we could use the movement of single molecules of FtsW as an indicator of its active state. We therefore first sought to define FtsW dynamics in Caulobacter. In E. coli, the PG synthases move with two dynamic modes: (1) fast (∼30 nm/s), inactive PG synthases driven by FtsZ treadmilling and (2) slow (∼8 nm/s), active PG synthases driven by PG synthesis (Yang et al., 2021). We sought to characterize the relationships between FtsWI activity, FtsZ dynamics, and FtsW dynamics in Caulobacter by performing SMT.
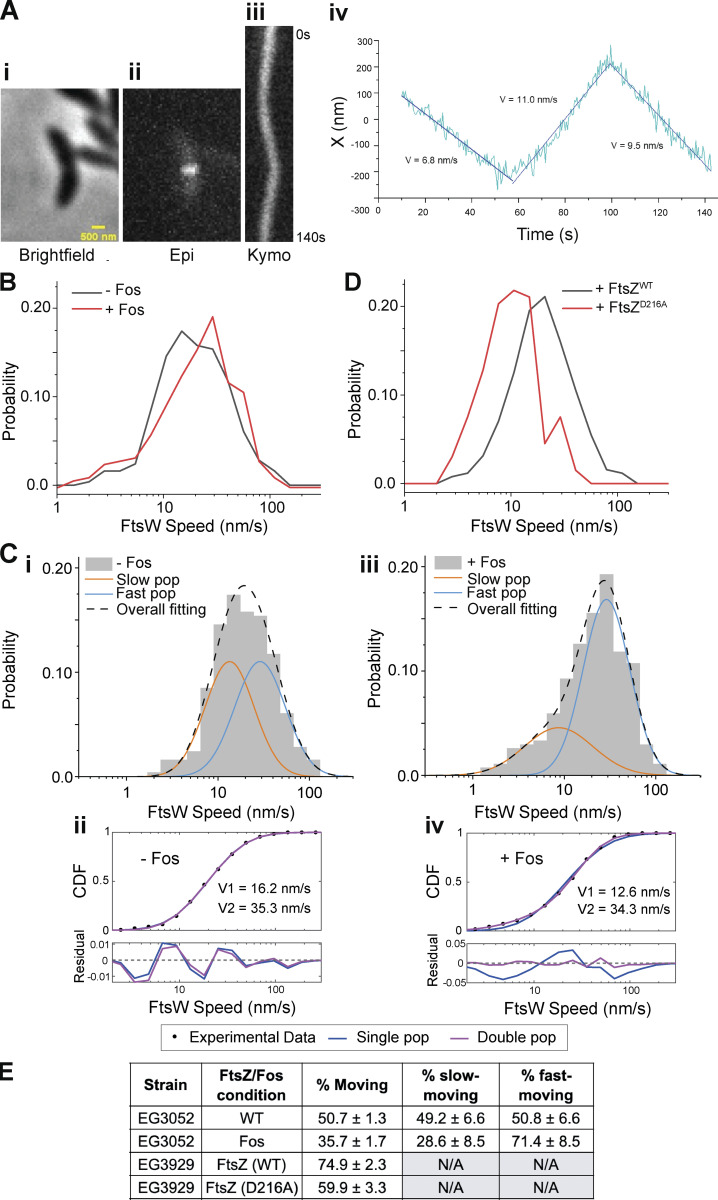
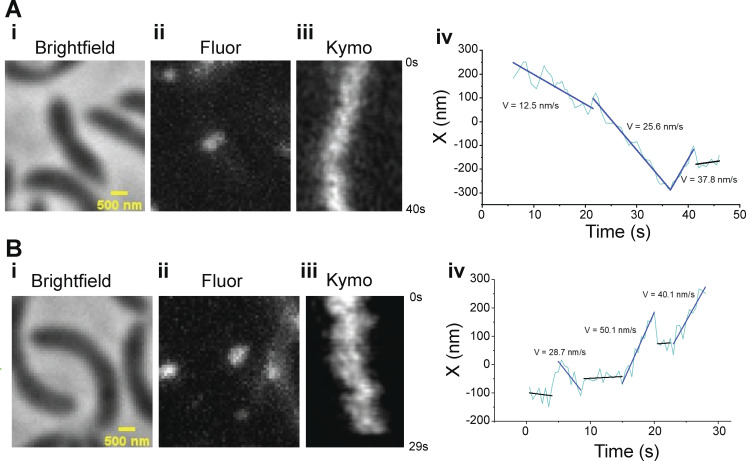
We generated a strain expressing halo-ftsW as the sole copy at the native locus in an otherwise WT background (EG3052). We labeled and measured single molecules of FtsW by titrating the levels of Janelia Fluor 646 Halo ligand (Grimm et al., 2017). While tracking FtsW, we observed single molecules that had processive movement, with some molecules changing speed, implying that FtsW can dynamically switch between movement modes, similar to observations in E. coli (Yang et al., 2021). To focus on FtsW molecules during division, we synchronized cells prior to analysis and imaged after ∼50 min of growth to ensure we were imaging predivisional and constricting cells. We measured speeds of mobile FtsW molecules at the division site by dividing the distances covered by the time to cover that distance (Fig. 2 A; Fig. S2, A and B; and Videos 2, 3, and 4). In some cases, a single molecule exhibited changes in the speed of movement, from fast to slow or slow to fast, and/or transitioned from moving to stationary or vice versa during measurement (Fig. S2, A and B; and Videos 3 and 4). Additionally, a proportion of molecules existed at the division plane without movement, which we define as the stationary population.
Figure 2.
FtsW in Caulobacter moves in two populations. (A i) Brightfield image of a Halo-ftsW**I* cell. (ii) Representative maximum fluorescence intensity projection (Epi) for Halo-FtsW**. (iii) Kymograph (Kymo) of the fluorescence signal of a line scan across the division plane for a Janelia Fluor 646–labeled single Halo-FtsW** (EG3053). (iv) Plot of molecule position at midcell over the course of imaging and speeds of movement for each segment. X indicates the short axis of the cell. (B) Speed distribution of directionally moving Halo-FtsW (EG3052) molecules with (n = 318 from four biological replicates) and without (n = 256 total from three biological replicates) Fos treatment. (C i and iii) Two-population fitting of the distributions in B without (i) or with (iii) Fos treatment. Orange and blue curves represent the slow- and fast-moving populations, respectively. The black dashed curve is the overall fit of the distribution. (ii and iv) Goodness of fit (top) of two- (magenta) versus one- (blue) population fitting of Halo-FtsW speed distribution. The residuals of each fit (bottom) illustrate how well each model captures the data at every point along the CDF. Data in C is duplicated in Fig. 3 B. (D) Speed distribution of directionally moving Halo-FtsW molecules in cells (EG3929) producing FtsZWT (n = 254 total from three biological replicates) or FtsZD216A (n = 131 total from three biological replicates). (E) Fraction of the FtsW population that is moving overall (% moving), slow moving, or fast moving for the indicated strains and conditions.
Figure S2.
Example trajectories of single molecules of Halo-FtsW showing FtsW may change speeds. (A and B i) Brightfield image of a halo-ftsW background cell (EG3052). (ii) Representative maximum fluorescence intensity projection image (Fluor) for single labeled Halo-FtsW. (iii) Kymograph (Kymo) of the fluorescence signal of a line scan across the division plane that encapsulates a labeled single molecule of Halo-FtsW**. (iv) Plot of the molecule position within the line scanning over the course of SMT.
Video 2.
FtsW** movement about the division plane in an ftsW**I* background. Representative single-molecule fluorescence time-lapse of fluorescent JF646-labeled Halo-FtsW** in an ftsW**I* background over 140 s. This time-lapse corresponds to the kymograph present in Fig. 2, A iii. Each frame is 500 ms.
Video 3.
FtsW moving at multiple speeds in a WT background. Representative single-molecule fluorescence time-lapse of fluorescent JF646-labeled Halo-FtsW in a WT background over 40 s. This time-lapse corresponds to the kymograph present in Fig. S2, A iii. Each frame is 500 ms.
Video 4.
FtsW moving at multiple speeds in a WT background. Representative single-molecule fluorescence time-lapse of fluorescent JF646-labeled Halo-FtsW in a WT background over 29 s. This time-lapse corresponds to the kymograph present in Fig. S2, B iii. Each frame is 500 ms.
To determine if FtsW movement is dependent on its activity, we inhibited lipid II synthesis using fosfomycin (Fos), thereby preventing FtsWI activity by depleting its substrate. In untreated cells, FtsW molecules moved at an average speed of 24.9 ± 1.1 nm/s (n = 318). Treatment with Fos caused an increase in average FtsW speed to 27.6 ± 1.2 nm/s (n = 256) and an increase in stationary molecules (50.7% moving in untreated and 35.7% moving in Fos-treated, Fig. 2 B and Table S1). These data are consistent with the two-population dynamics model from E. coli wherein inactive FtsW molecules are stationary or move at a faster speed than active FtsW. To determine if Caulobacter FtsW exhibits two-population dynamics, we compared one- and two-population fitting of the cumulative probability density function (CDF) of untreated FtsW SMT data (see Materials and methods). While the two-population fitting identified fast- and slow-moving populations with average velocities at ∼16 and ∼35 nm/s, respectively, the one-population model fit the data equally well (Fig. 2, C i and ii). We reasoned that this may be because, in unperturbed conditions, the two populations are roughly equal in proportion and have overlapping velocity distributions. We hypothesized that when PG synthesis is inhibited by Fos, the inactive population would be more prevalent since FtsW is no longer able to synthesize PG. Indeed, when Fos is present, two-population modeling of FtsW dynamics fit the data best and there was a larger proportion of fast-moving molecules compared with that in the absence of Fos (71.4% ± 8.5% fast-moving +Fos versus 50.8% ± 6.6% fast-moving −Fos, Fig. 2, C iii and iv and E; and Table S1). The average velocities of the fast- and slow-moving populations were 34.3 and 12.6 nm/s, respectively (Fig. 2, C iii and Table S1). These data together suggest that inactive FtsW molecules are either stationary or move faster on average than active FtsW molecules. In subsequent two-population modeling, we fixed the velocity of the fast-moving population to isolate effects on the slow-moving population.
In E. coli, movement of the fast-moving population of FtsW is driven by FtsZ treadmilling. The fast-moving population of FtsW in untreated WT Caulobacter has an average velocity of 35.3 nm/s (Fig. 2, C ii and Table S1), similar to the velocity of fast-moving PG synthases and of FtsZ treadmilling (∼30 nm/s) in E. coli. Due to the small size of Caulobacter we were unable to resolve FtsZ clusters sufficiently to determine their treadmilling speeds. Instead, we employed a GTPase-deficient variant of FtsZ (FtsZD216A; Barrows et al., 2023, Preprint) to perturb FtsZ dynamics in vivo and test the hypothesis that FtsZ dynamics impact the movement of FtsW. We generated a strain producing Halo-FtsW in which WT ftsZ expression is driven by vanillate and ftsZD216A expression is induced by xylose. We removed vanillate and depleted WT FtsZ for 1 h, synchronized cells to isolate swarmers, and then induced ftsZWT or ftsZD216A with vanillate or xylose, respectively, for 1 h prior to SMT of Halo-FtsW. We found that FtsW molecules in cells producing FtsZWT moved at an average velocity of 24.5 ± 1.1 (n = 254; Fig. 2 D and Table S1). When FtsZD216A was produced instead, the average velocity of FtsW was dramatically slower (12.2 ± 0.7 nm/s, n = 131; Fig. 2 D and Table S1). This observation is consistent with FtsZ dynamics driving movement of a fast population of FtsW molecules. When FtsZ dynamics are slowed by abrogating GTP hydrolysis, FtsZ-associated FtsW molecules move at a slower rate that is indistinguishable from that of slow, active FtsW molecules.
Hyperactive Halo-FtsW molecules move more slowly on average than WT Halo-FtsW
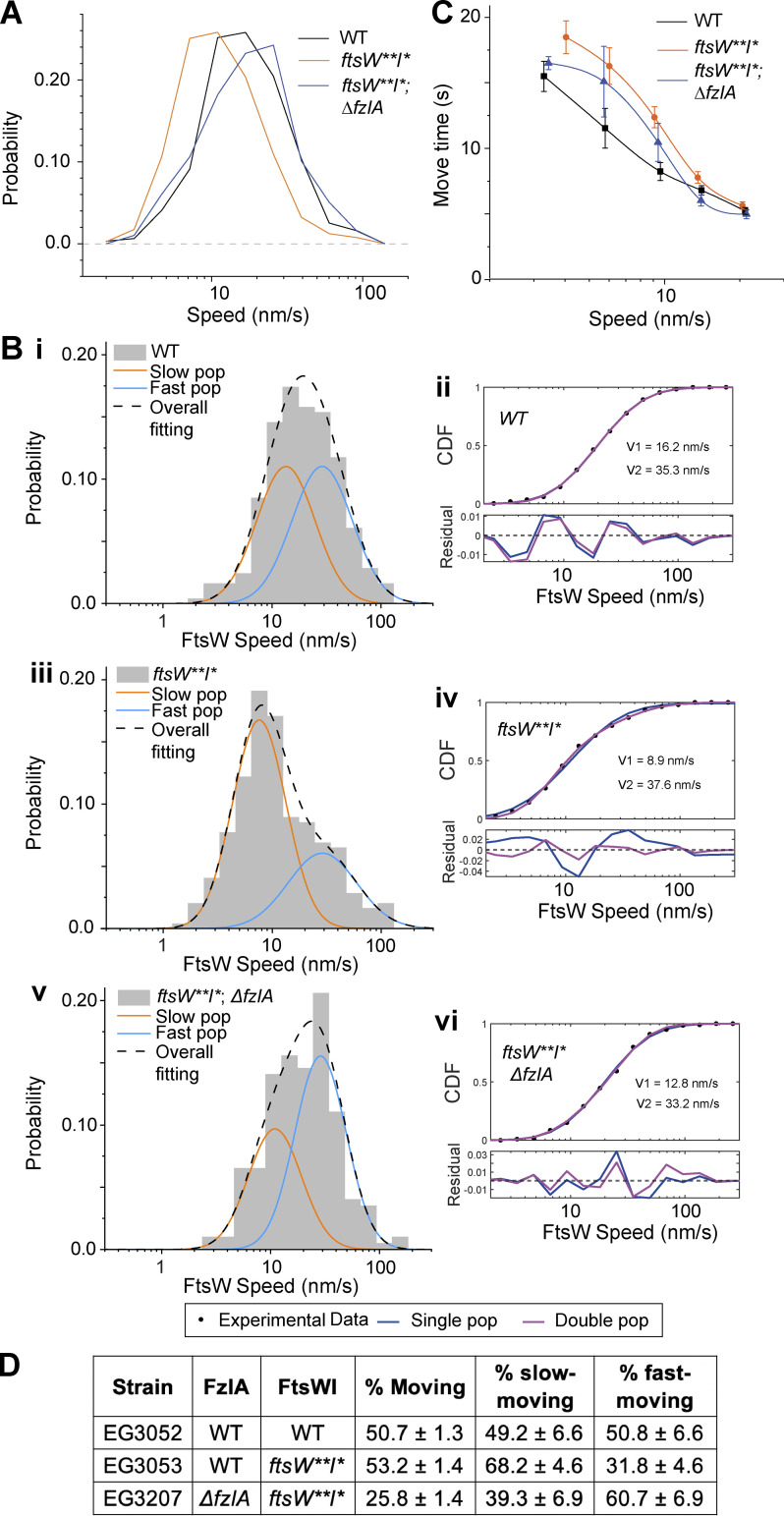
Having established the effects of perturbing PG synthesis and FtsZ dynamics on FtsW movement in Caulobacter, we next sought to determine how activating mutations in FtsWI impact FtsW dynamics. To do this, we generated a Halo-FtsW** fusion in an ftsW**I* hyperactive background and performed SMT of Halo-FtsW**. We observed that FtsW** molecules move more slowly on average (16.6 ± 0.7 nm/s, n = 403) than WT FtsW (24.9 ± 1.1 nm/s, n = 318; Fig. 3 A and Table S1). By comparing the two-population fitting for WT and ftsW**I* backgrounds, we observed that FtsW** has a larger slow-moving population (68.2 ± 4.6%) than WT FtsW (49.2 ± 6.6%; Fig. 3, B i–iv and Table S1).
Figure 3.
Mutations to FtsW or FzlA impact FtsW movement. (A) Histogram comparing single-molecule speeds of FtsW or FtsW** in halo-ftsW (WT; EG3052, n = 318 total from four biological replicates), halo-ftsW**; ftsI* (ftsW**I*; EG3053, n = 403 total from three biological replicates), or ΔfzlA; halo-ftsW**; ftsI* (ΔfzlA, ftsW**I*; EG3207, n = 198 total from three biological replicates) backgrounds. (B i, iii, and v) Two-population fitting of the histograms of A ((i) FtsW in WT, (iii) FtsW** in ftsW**I*, (v) FtsW** in ΔfzlA, ftsW**I*). Orange and blue curves represent the slow- and fast-moving populations, respectively. The black dashed curve is the overall fit of the distribution. (ii, iv, and vi) Goodness of fit (top) of two- (magenta) versus one- (blue) population fitting of Halo-FtsW speed distribution. The residuals of each fit (bottom) illustrate how well each model captures the data at every point along the CDF. Data in Fig. 2 C is duplicated in B. (C) Plot comparing the move time of single FtsW or FtsW** molecules at speeds <20 nm/s in halo-ftsW (WT; EG3052, n = 318 total from four biological replicates), halo-ftsW**; ftsI* (ftsW**I*; EG3053, n = 403 total from three biological replicates), or ΔfzlA; halo-ftsW**, ftsI* (ΔfzlA, ftsW**I*; EG3207, n = 198 total from three biological replicates) backgrounds. Error bars are SEM. (D) Fraction of the FtsW population that is moving overall (% moving), slow moving, or fast moving for the indicated strains and conditions.
To compare the processivity of mobile FtsW molecules between the ftsW**I* and WT backgrounds, we plotted the speeds of moving molecules against the time those molecules spent moving. To enrich for active FtsW for this analysis, we excluded molecules moving faster than 20 nm/s. We selected this value as ∼20 nm/s, which was the point at which fast-moving molecules became more abundant than slow-moving molecules in the WT two-population modeling. We found that slow-moving FtsW** molecules travel for longer periods of time than slow-moving WT FtsW on average (Fig. 3 C). Since slow-moving PG synthases are more likely to be active, these data suggest that active FtsW** molecules are more processive than active WT FtsW molecules. Note that the average velocity of the slow-moving population of FtsW** (8.9 ± 0.6 nm/s) is slower than that of the slow-moving population of WT FtsW (16.2 ± 1.5 nm/s; Fig. 3, B i–iv). This could be due to differences in PG synthetic rates of WT versus FtsW**I* and/or to better precision in the velocity measurement of FtsW** as more data points were available due to high processivity.
Interestingly, we did not observe a change in the percentage of stationary FtsW molecules when FtsW was hyperactivated (roughly 50% in both WT and ftsW**I*, Fig. 3 D). In E. coli, stationary FtsW molecules are not active for cell wall synthesis but may be poised for activation (Yang et al., 2021), and we propose that stationary FtsW molecules in Caulobacter behave similarly. Collectively, our data indicate that ftsW**I* cells constrict faster due to both a greater proportion of active (slow-moving) PG synthase complexes and cytokinetic PG synthases polymerizing for longer stretches than WT complexes. These data support a model where FtsW dynamics in Caulobacter are similar to those in E. coli, with a slow-moving, active population driven by PG synthesis and a fast-moving, inactive population driven by FtsZ.
Absence of FzlA decreases the proportion of active Halo-FtsW** in an ftsW**I* background
With an experimental framework in hand to understand the relationship between FtsW movement and activity, we next addressed the role of FzlA in regulating FtsW activity. We deleted fzlA in the ftsW**I* background producing Halo-FtsW** and replaced it with a gentamycin resistance cassette at the fzlA locus (Fig. S1 C). Notably, in the absence of FzlA (EG3207), moving FtsW** molecules (26.1 ± 1.6 nm/s, n = 198) had average speeds similar to FtsW in a WT background (24.9 ± 1.1 nm/s, n = 318) and faster than FtsW** in the presence of FzlA (16.6 ± 0.7, n = 403; Table S1 and Fig. 3 A). However, the proportion of inactive, stationary PG synthases increased when compared with either FtsW** or FtsW in the presence of FzlA (Table S1 and Fig. 3 D, 75% in ΔfzlA versus ∼50% in WT or ftsW**I*).
To clarify the impacts of FzlA on the activation state of FtsW, we modeled two-population dynamics in the ΔfzlA background, which fit the distribution better than a single population (Fig. 3, B ii and iv). Deletion of fzlA resulted in a greater proportion of fast-moving, inactive FtsW** (60.7% ± 6.9%) when compared with the hyperactive strain with fzlA (31.8% ± 4.6%) or the WT strain (50.8% ± 6.6%; Fig. 3, B i–vi and D; and Table S1). The effects of FzlA on FtsW activity are consistent with the cell length and constriction rate defects associated with fzlA deletion we previously reported (Lariviere et al., 2019). These data also indicate that FtsW**I* can still receive activating signals from FzlA even though fzlA is no longer essential. FtsW** in the ΔfzlA background had processivity and moving velocity similar to FtsW** in the presence of FzlA (Fig. 3 C), suggesting that FzlA does not affect the activity of individual PG synthase molecules, but rather the fraction of the population that is active (Fig. 3 D).
Depletion of FzlA in WT cells decreases the active, slow-moving FtsW population
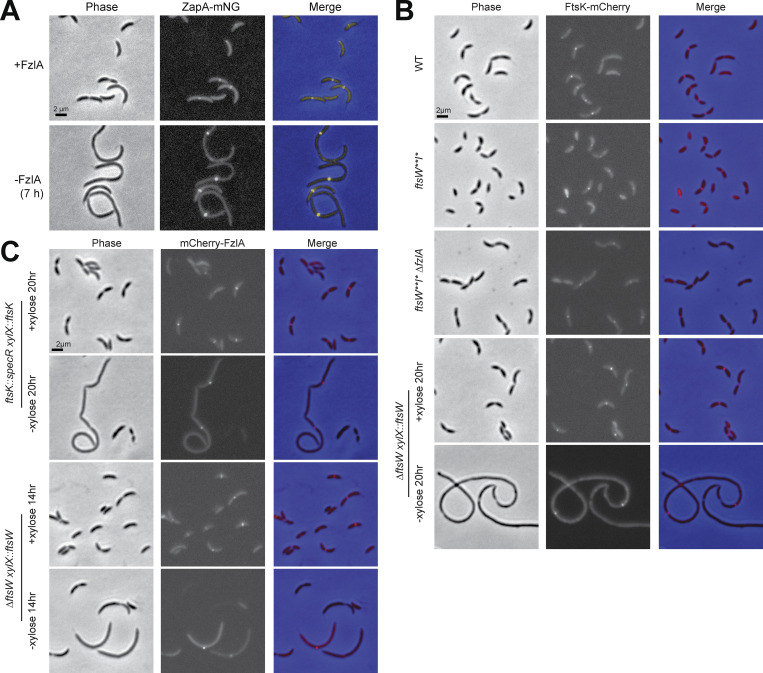
To examine the effects of loss of FzlA on FtsW dynamics in a WT background, we generated a strain-dependent on xylose for fzlA expression with natively expressed WT halo-ftsW (EG3523). When xylose inducer was removed, FzlA was undetectable after 7 h and cells began to filament (Fig. S1, D and E; and Fig. S3 A). To ensure measurement of FtsW molecules associated with Z-rings in these filamentous cells, we replaced zapA at the native locus with zapA-mNeonGreen (zapA-mNG; Fig. S3 A). ZapA is an FtsZ-binding protein that reliably reports on Z-ring positioning (Woldemeskel et al., 2017).
Figure S3.
ZapA localization reflects Z-ring position in filamentous cells depleted of FzlA, and FtsK and FzlA localize independently of each other and of FtsW. (A) Phase-contrast, epifluorescence, and merged images of a strain expressing zapA-mNG dependent on xylose for production of FzlA (EG3523). The strain was either continuously induced (top) or depleted for 7 h (bottom) by being grown in the presence of 0.001% xylose or glucose, respectively. (B) Representative phase-contrast, epifluorescence, and merged images of ftsk-mCherry at its native locus in a WT (EG1291), ftsW**I* (EG3926), or ftsW**I* ∆fzlA (EG2481) background, and in an FtsW depletion strain (EG740). The FtsW depletion strain was grown in the presence (+FtsW) or absence (−FtsW) of 0.3% xylose for 20 h and then imaged. (C) Representative phase-contrast, epifluorescence, and merged images of vanillate inducible mCherry-fzlA in either an FtsKFL (EG3935) or FtsW (EG3928) depletion strain. The FtsKFL depletion strain was grown in the presence (+FtsKFL) or absence (−FtsKFL) of 0.3% xylose for 20 h before imaging. The FtsW depletion strain was grown in the presence (+FtsW) or absence (−FtsW) of 0.3% xylose for 14 h before imaging. mCherry-fzlA production was induced in both strains by adding 0.5 mM of vanillate for 2 h before imaging. Scale bars are all 2 µm.
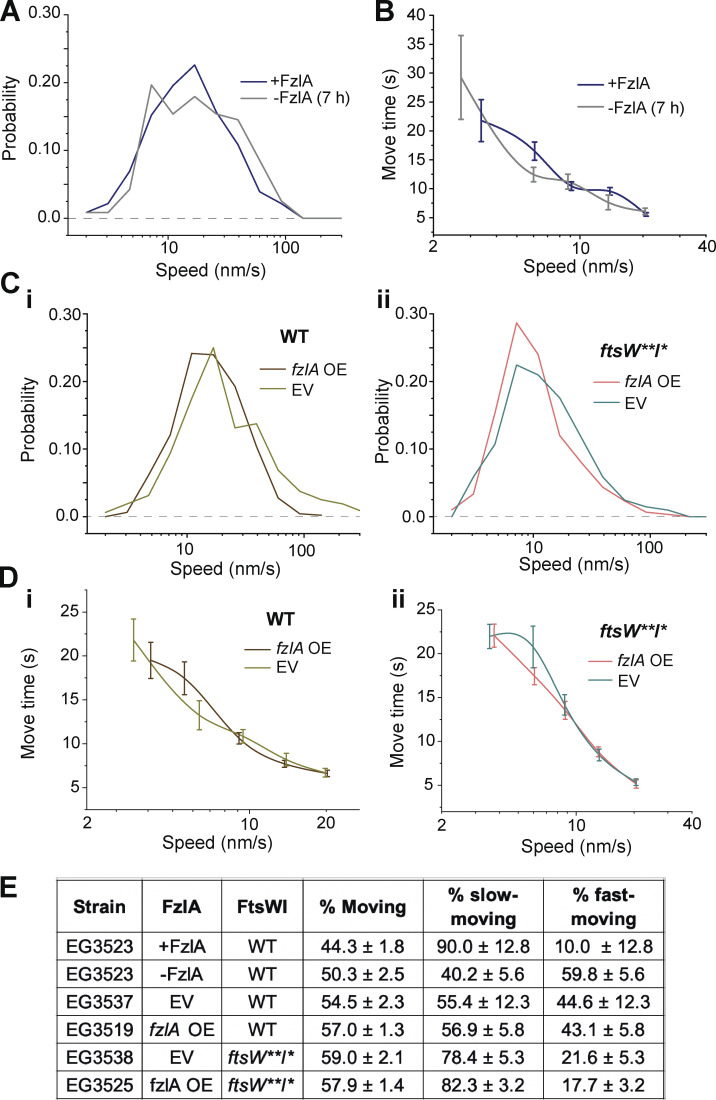
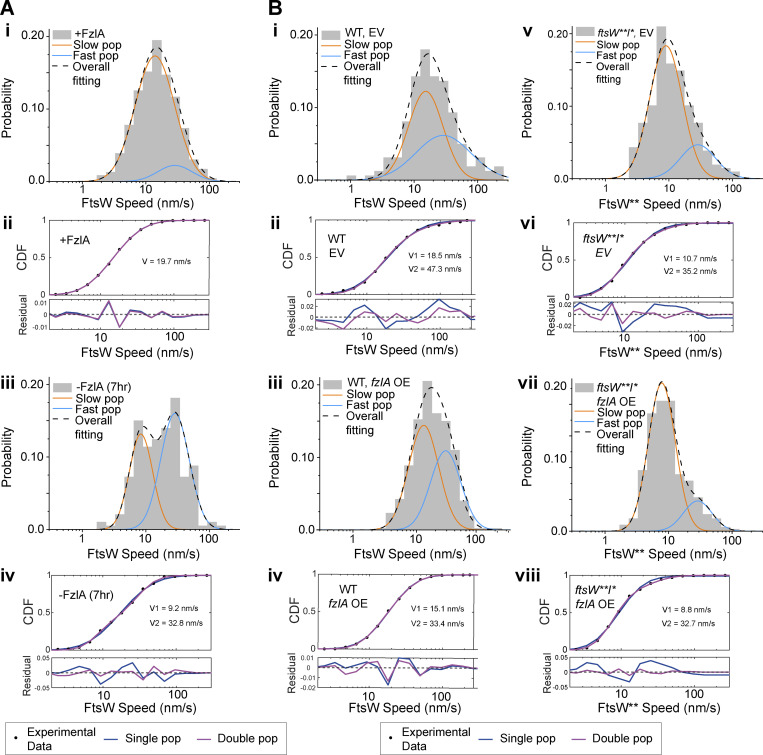
When comparing FtsW dynamics between induced and depleted conditions, we observed that overall FtsW moves faster on average when FzlA is absent (Fig. 4 A, 23.0 ± 1.9 nm/s without FzlA, n = 117 versus 19.6 ± 1.1 nm/s with FzlA, n = 230, Table S1), consistent with our ∆fzlA results and suggesting fewer active PG synthase complexes are present without FzlA. FtsW processivity was not affected by FzlA in this background (Fig. 4 B), similar to our observations using the ftsW**I* strain with and without FzlA. FzlA-replete conditions were modeled equally well by one- or two-population dynamics, but FzlA depletion was modeled significantly better by two-population dynamics, with more than half the molecules present being in the fast-moving population (Fig. 4 E; Fig. S4, A i and ii; and Table S1). These data are consistent with FzlA-mediated signaling acting to convert FtsW into an activated, slow-moving state.
Figure 4.
Depletion or overproduction of FzlA impacts FtsW movement. (A) Histogram comparing single-molecule speeds of FtsW in cells (EG3523) producing FzlA (+FzlA, 0.001% xylose, n = 230 total from three biological replicates) or depleted of FzlA for 7 h (−FzlA [7 h], 0.001% glucose, n = 117 total from three biological replicates). (B) Plot comparing the move time of single FtsW molecules moving at <20 nm/s in cells producing FzlA (+FzlA, 0.001% xylose, n = 230 total from three biological replicates) or depleted of FzlA for 7 h (−FzlA [7 h], 0.001% glucose, n = 117 total from three biological replicates). Error bars are SEM. (C i and ii) Histogram comparing speeds of FtsW or FtsW** in cells with an EV control or with an fzlA overexpression (OE) construct in either (i) a halo-ftsW (EV; EG3537, n = 472 total from three biological replicates. fzlA OE; EG3519, n = 300 total from three biological replicates) or (ii) halo-ftsW**; ftsI* (EV; EG3538, n = 160 total from four biological replicates. fzlA OE; EG3525, n = 205 total from three biological replicates) background. (D i and ii) Plot comparing the move time of single FtsW and FtsW** molecules moving at speeds <20 nm/s with and without FzlA overproduction in a (i) halo-ftsW or (ii) halo-ftsW**; ftsI* background. Error bars are SEM. (E) Fraction of the population that is moving overall (% moving), slow moving, or fast moving for the indicated strains and conditions.
Figure S4.
FzlA converts FtsW into a slow-moving, active state. Two-population fitting of the histograms of FtsW-HaloTag or FtsW**-HaloTag molecules under different conditions. The orange and blue curves represent the slow- and fast-moving populations, respectively. The black dashed curve is the overall fitting profile of the distribution. (A i and iii) (i) FtsW in FzlA-producing cells (EG3523 + 0.001% xylose); (iii) FtsW in FzlA-depleted cells (EG3523 + 0.001% glucose). (B i, iii, v, and vii) (i) FtsW in EV control cells; (iii) FtsW in FzlA-overproducing cells; (v) FtsW** in ftsW**I* EV control cells; (vii) FtsW** in ftsW**I* FzlA-overproducing cells. (A ii and iv; and B ii, iv, vi, and viii) The fit goodness of one- (blue) and two- (magenta) population fitting of the corresponding conditions. The top sub-figures are the CDF curves and the fit curves, while the bottom figures show the residuals of the fitting.
We next assessed the stationary FtsW population in our depletion strain with and without FzlA. Surprisingly, depletion of FzlA in a WT background did not change the proportion of total moving FtsW molecules (Fig. 4 E and Table S1, ∼50% in both deplete and induced), unlike in the ΔfzlA background (Fig. 3 D and Table S1, 25%). To clarify this difference, we sequenced the genomes of the FzlA depletion and ∆fzlA strains used for FtsW SMT. We found that the ∆fzlA strain (EG3207) carried two point mutations (introducing a premature stop in ubiB and a T338A mutation in divL [Table S4]) not present in the depletion strain, which could be responsible for this difference in effect on FtsW dynamics. Although DivL indirectly regulates division through effects on cell cycle master regulators (van Teeseling and Thanbichler, 2020), neither DivL nor UbiB have been implicated in cell wall metabolism or cell division directly. Alternatively, FtsW** may have an increased affinity for a stationary target compared with WT FtsW, which is counteracted by the loss of FzlA.
Slow-moving, active FtsW molecules are more abundant when FzlA is overproduced
Having assessed the effects of loss of FzlA on FtsWI dynamics and activity, we next turned to cells overproducing FzlA. We initiated this study with the observation that FzlA overproduction hyperactivates constriction. To identify changes to PG synthase activation during FzlA overproduction, we compared Halo-FtsW speeds from strains with the fzlA overexpression construct (EG3519) or EV (EG3537) in the presence of 0.3% xylose to induce fzlA overexpression for 1 h prior to synchrony and over the course of the experiment (Fig. S1 F). With FzlA overproduction, the average FtsW speed was reduced compared with EV (Fig. 4, C i and Table S1, 23.2 ± 0.9 nm/s with FzlA overproduction, n = 300 versus 32.3 ± 3.6 nm/s for the EV control, n = 472). Additional FzlA altered neither the processivity nor the proportion of moving molecules (Fig. 4, D i and E; and Table S1). These results reinforce our conclusion that FzlA-mediated signaling converts FtsW to an active state.
Our prior results demonstrated that FzlA-mediated signaling still impacts FtsW**, which suggests that FzlA-mediated hyperactivation can occur in an ftsW**I* background, as well. In line with this idea, we observed that FzlA overproduction in this background (EG3525) further stimulated FtsW**, as speeds decreased to the slowest observed average speed when FzlA was overproduced compared with EV (EG3538; Fig. 4, C ii and Table S1, 13.5 ± 0.8 nm/s for overproduced FzlA, n = 205 versus 16.3 ± 1.2 nm/s for EV, n = 160). There was no change to processivity or to the proportion of moving PG synthases when FzlA was overproduced (Fig. 4, D ii and E; and Table S1). In both the WT and ftsW**I* backgrounds, FzlA overproduction resulted in higher proportions of slow-moving FtsW (Fig. 4 E; Fig. S4, B i–viii; and Table S1). These results further demonstrate that FzlA provides activating signals to the PG synthases, including FtsW**I*, in which the complex is already hyperactive.
FzlA can move with active FtsWI
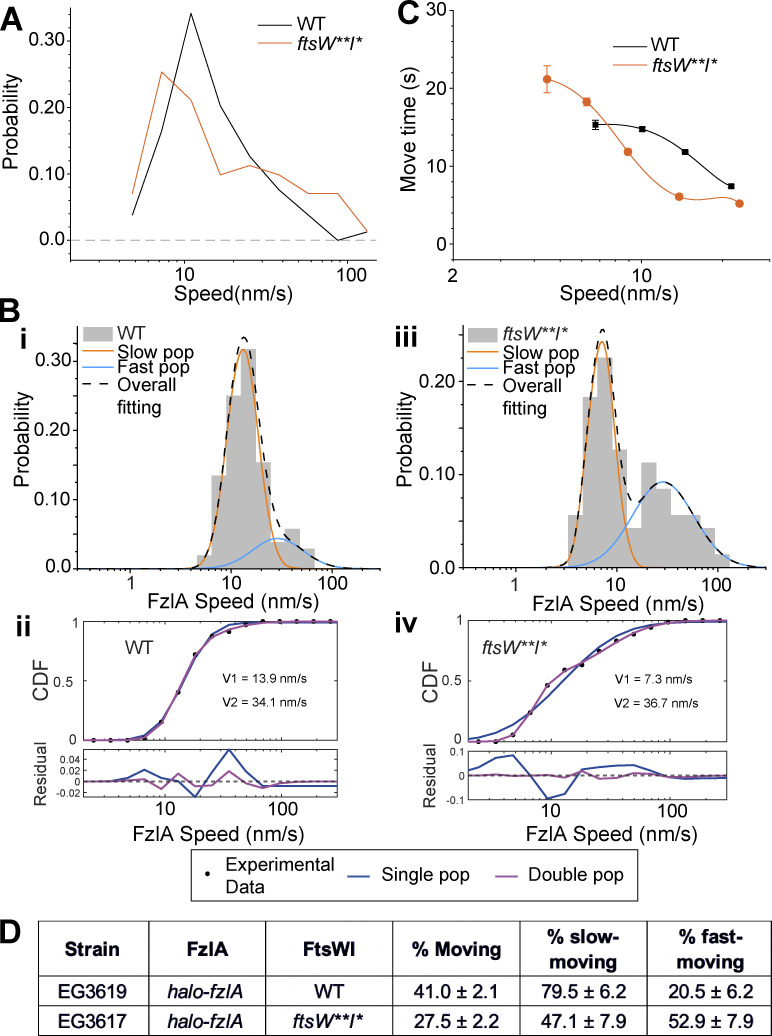
Considering that FzlA participates in FtsWI activation, we sought to test whether FzlA remains associated with activated FtsWI or dissociates after signaling. To do this, we assessed FzlA dynamics using SMT. We constructed strains expressing halo-fzlA at the native fzlA locus in WT (EG3619) and ftsW**I* (EG3617) backgrounds. Like Halo-FtsW, we observed processive movement of Halo-FzlA at the division site, as well as stationary molecules (41.0% ± 2.1% versus 27.5% ± 2.2% moving in WT and ftsW**I* backgrounds, respectively, Table S1). Average FzlA speeds were similar in the WT and ftsW**I* backgrounds (Fig. 5 A and Table S1, 17.3 ± 1.1 nm/s, n = 104 and 20.3 ± 2.5 nm/s, n = 71, respectively). However, the distribution of Halo-FzlA speeds was broader in an ftsW**I* background than in WT, with more very slow- and very fast-moving molecules than in WT (Fig. 5 A). This was also evident when the Halo-FzlA speeds in both the WT and ftsW**I* backgrounds were modeled using two-population dynamics, which fit the results significantly better than one-population modeling (Fig. 5, B i–iv and Table S1). The average FzlA speed for the slow population was slower in an ftsW**I* background (7.7 ± 0.5 nm/s) than in WT cells (12.4 ± 0.7 nm/s; Fig. 5, B i–iv and Table S1), similar to what we observed for FtsW** (8.9 ± 0.6 nm/s) compared with WT FtsW (16.2 ± 1.5 nm/s; Fig. 3, B i–iv and Table S1). We propose that fast-moving FzlA molecules may be traveling with FtsZ cluster ends, consistent with our earlier results that fast-moving FtsW depends on FtsZ dynamics (Fig. 2 D) and the fact that FzlA is dimeric and can bind directly to FtsZ (Goley et al., 2010). The presence of slow-moving FzlA molecules with speeds similar to those of FtsW suggests that they can remain associated with activated FtsWI complexes as they synthesize PG. Interestingly, there was a higher percentage of the fast-moving FzlA population in the ftsW**I* background. This is likely because active FtsW**I* does not require FzlA for activity, resulting in FzlA remaining stationary more often in ftsW**I* (∼72%) than in WT cells (∼59%; Fig. 5, B i–iv and D; and Table S1). Stationary FzlA molecules are likely bound to the interior of FtsZ filaments and treadmilling with FtsZ. Finally, FzlA was less processive in the FtsW**I* background than in WT at speeds of 10–20 nm/s, but this difference diminished at slower FzlA speeds (Fig. 5 C). We conclude that FzlA can remain associated with active, slow-moving FtsWI complexes.
Figure 5.
FzlA moves with active FtsWI. (A) Histogram comparing speeds of FzlA molecules in halo-fzlA; WT (EG3619, n = 104 total from three biological replicates) or halo-fzlA; ftsW**I* (EG3617, n = 71 total from four biological replicates) backgrounds. (B) Two-population fitting of FzlA single molecule movement data from A. (i and iii) Two-population fit to FzlA single molecule movement in WT (EG3619) (i) and ftsW**I* (EG3617) (iii). Orange and blue curves represent the slow- and fast-moving populations, respectively. The black dashed curve is the overall fit of the distribution. (ii and iv) Goodness of fit (top) of two- (magenta) versus one- (blue) population fitting of halo-fzlA speed distribution. The residuals of each fit (bottom) illustrate how well each model captures the data at every point along the CDF. (C) Plot comparing the move times of FzlA molecules moving at speeds <20 nm/s in halo-fzlA; WT (EG3619, n = 93 total from three biological replicates) or halo-fzlA; ftsW**I* (EG3617, n = 55 total from four biological replicates) backgrounds. Error bars are SEM. (D) Fraction of the FzlA population that is moving overall (% moving), slow moving, or fast moving for the indicated strains and conditions.
FzlA overproduction causes lethal division events
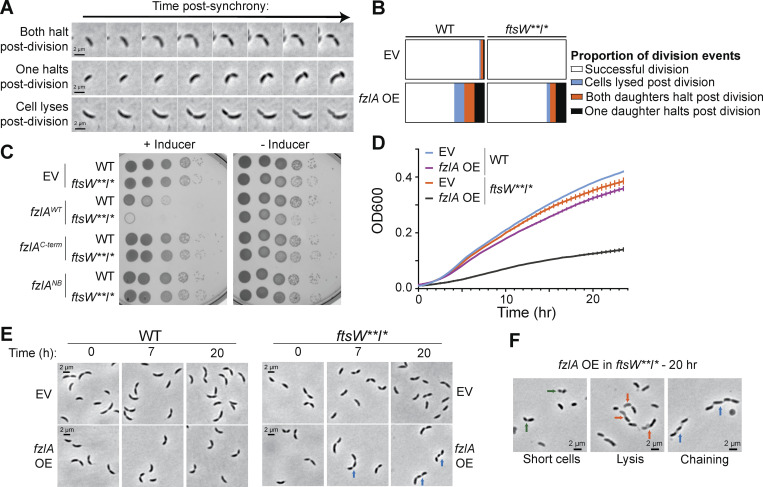
Previous work demonstrated that hyperactivating FtsWI affects cell wall and envelope integrity (Modell et al., 2014; Lariviere et al., 2019). These observations led us to ask if there are phenotypic consequences of FtsWI hyperactivation through overproduction of FzlA. While analyzing time-lapse of fzlA-overexpressing WT cells, we noticed that cells sometimes died during or right after division. The lethal division events we observed fell into three major categories; (1) both daughters halt growth, (2) one daughter halts growth, and (3) one daughter lyses (Fig. 6, A and B; and Videos 5, 6, and 7). These lethal division events were rare in the EV control strain, suggesting that hyperconstriction caused by excess FzlA is deleterious to cells.
Figure 6.
FzlA overproduction is toxic. (A) Representative phase-contrast time-lapse images of constricting cells that result in lethal division events due to FzlA overproduction. (B) Quantification of lethal divisions in the WT and ftsW**I* background comparing EV controls (WT: EG1644, n = 182 division events; ftsW**I*: EG3466, n = 145) and to fzlA overexpression (OE; WT: EG3637, n = 155; ftsW**I*: EG3467, n = 140). (C) Spot dilutions of WT or ftsW**I* strains harboring an EV control or fzlA variant overexpression construct, plated with 0.3% xylose (+Inducer) or 0.2% glucose (−Inducer). (D) Growth curves of WT or ftsW**I* strains harboring an EV control or fzlA-overexpression construct. Strains were pre-induced for 6 h with 0.3% xylose and induction continued with 0.3% xylose during the experiment. Points are the mean of three technical replicates at that timepoint, error bars are SEM. Shown is a representative replicate of a biological triplicate. (E) Phase-contrast time course of WT or ftsW**I* cells harboring an EV control or fzlA-overexpression construct. (F) Example phase-contrast images of cells after 20 h of FzlA overproduction in an ftsW**I* background. Arrow colors: green = very short cells, orange = one half of the constricted cell lysed, blue = cell chaining.
Video 5.
A division in which both daughter cells halt growth after constriction. Representative phase-contrast microscopy time-lapse of a FzlA-overproducing cell in a WT background for which both daughter cells halt growth after division. Each frame is 5 min apart and the total time-lapse is 4 h.
Video 6.
A division in which one daughter cell halts growth after constriction. Representative phase-contrast microscopy timelapse of a FzlA-overproducing cell in a WT background for which one daughter cell halts growth after division. Each frame is 5 min apart and the total time-lapse is 4 h.
Video 7.
A division in which one daughter cell lyses growth after constriction. Representative phase-contrast microscopy timelapse of a FzlA overproducing cell in a WT background for which one daughter cell lyses after division. Each frame is 5 min apart and the total time-lapse is 4 h.
Interestingly, without FzlA overproduction in an ftsW**I* background (EG3466), we did not observe lethal divisions, suggesting hyperactive PG synthases are not sufficient to cause an increase in lethal division events on this timescale. FzlA overproduction in ftsW**I* (EG3467) increased lethal division events compared with the EV control but not greater than that in a WT background (Fig. 6 B). To understand how division failures affect long-term viability, we performed growth and viability analyses of WT and ftsW**I* cells with and without FzlA overproduction. Overexpression of fzlA in a WT background caused a 2-log reduction in colony-forming units and a moderate reduction in doubling time in liquid media (Fig. 6, C and D). Strikingly, long-term overexpression of fzlA was lethal in an ftsW**I* background, both on solid and liquid media (Fig. 6, C and D).
FzlA has two essential surfaces: an FtsZ-binding surface and a C-terminal tail of unknown function (Lariviere et al., 2018). We sought to test whether either function is required for the toxicity associated with FzlA overproduction. To do this, we assessed the effects of overexpression of mutant fzlA with disrupted FtsZ-binding (fzlANB2) or with a lethal charge-reversal in the C-terminal tail (fzlAD227K) in the WT or ftsW**I* backgrounds. Neither fzlAD227K nor fzlANB2 overexpression was lethal in either background, demonstrating that FzlA requires both essential activities for toxicity (Fig. 6 C; and Fig. S1, A and B). The observation that fzlAD227K, which binds FtsZ similarly to WT but is unable to promote constriction (Lariviere et al., 2018), is not lethal when overproduced, suggests that the detrimental effects of FzlA overproduction do not occur through FzlA-mediated modulation of FtsZ assembly.
To better understand how FzlA overproduction in an ftsW**I* background causes toxicity, we imaged cells after 20 h of FzlA overproduction. We observed cells that lysed during division (red arrows), as well as cells that were significantly shorter (green arrows) than ftsW**I* cells with an EV (Fig. 6, E and F). Interestingly, we also observed cell chaining (blue arrows). The cells that chained had multiple deep constrictions with cell bodies remaining connected, suggesting that division failed to complete at a late stage and then initiated again at a second site.
FtsK is a potential binding partner of FzlA
Cell chaining has been observed in cells with mutations to cell wall hydrolases, outer-membrane components, and factors that regulate genome integrity (Meier et al., 2017; Bernhardt and de Boer, 2004; Peters et al., 2011; Heidrich et al., 2001; Wang et al., 2006; Uehara et al., 2010; Lo Sciuto et al., 2014). To better understand both the mechanism for the cell division defects that result from FzlA overproduction and how FzlA signals to activate FtsWI, we sought to identify downstream interactors of FzlA. To do this, we performed coimmunoprecipitation (coIP) in a strain (EG2217) that carries 3x-FLAG-fzlA as the sole copy at the native locus and subjected the eluate to mass spectrometry (MS)–based protein identification using WT cells as a negative control. MS identified FtsZ, a known interactor for FzlA, in the 3xFLAG-FzlA eluate, validating this approach (Table S2). From the proteins identified and enriched more than fivefold in the 3xFLAG-FzlA eluate, we identified FtsK, a cell division protein, as a putative binding partner (Table S2). Notably, neither FtsW nor FtsI was identified by MS, which indicates FzlA-mediated activation is indirect.
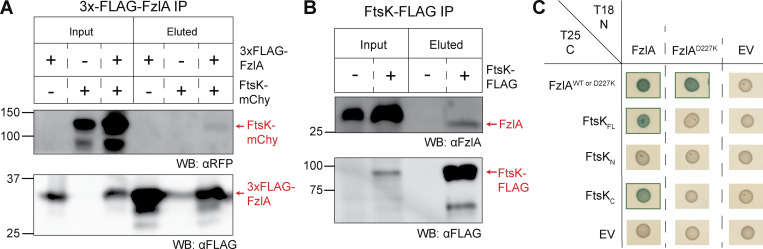
FtsK is an essential division protein with an N-terminal domain (FtsKN: residues 1–213) containing four transmembrane passes and a cytoplasmic C-terminal domain (FtsKC: residues 315–825), connected by a disordered linker (residues 213–315; Wang et al., 2006). FtsKN is responsible for localizing FtsK to the divisome and plays an essential role in constriction, while FtsKC is a hexameric DNA translocase that segregates the chromosomal termini away from the division site. Both domains are essential in Caulobacter (Wang et al., 2006). To confirm our MS results, we generated a strain in which ftsK-mChy was expressed at the ftsK locus as the sole copy in WT (EG2427) and 3xFLAG-fzlA (EG2428) backgrounds and performed anti-FLAG coIP followed by immunoblotting. Western blotting revealed that 3xFLAG-FzlA interacted with FtsK-mChy in vivo (Fig. 7 A). Conversely, we performed reciprocal coIP in a strain that harbored ftsK-FLAG at the ftsK locus as the sole copy (EG742) and compared it with WT. Western blot analysis demonstrated that FtsK-FLAG interacts with native, untagged FzlA in vivo as well (Fig. 7 B).
Figure 7.
FzlA interacts with FtsKC via the FzlA C-terminal tail. (A and B) Western blots of whole cell lysates or eluates from an immunoprecipitation (IP) using α-FLAG resin. Ladder values to the left of the blots are in kDa. Red arrows to the right of the blots indicate to what protein the band is attributed. The presence or absence of the fusion proteins in the strains used are indicated above the blots by plus (+) or minus (−) signs, respectively. (A) 3xFLAG-FzlA immunoprecipitation. Blots were incubated with primary antibodies recognizing either mCherry (αRFP) or FLAG (αFLAG). Lane 1 and 4, fzlA::3xFLAG-fzlA (EG2217). Lane 2 and 5, ftsK:: ftsK-mChy (EG2427). Lane 3 and 6, fzlA::3xFLAG-fzlA; ftsK:: ftsK-mChy (EG2428). (B) FtsK-FLAG immunoprecipitation. Blots were incubated with primary antibodies recognizing either FzlA (αFzlA) or FLAG (αFLAG). Lane 1 and 3, WT (EG865). Lane 2 and 4, ftsK::ftsK-FLAG (EG742). (C) BTH results for interaction between FzlA variants and full-length FtsK or its domains. The adenylyl cyclase subunits T18 (left) and T25 (top) are fused to proteins via the N-terminus or C-terminus, respectively. A green box around the representative spot image indicates that the three biological triplicates were positive for induction of the cAMP-dependent β-galactosidase reporter, indicating a positive interaction. Some data in C is duplicated in Fig. S5 A. Source data are available for this figure: SourceData F7.
The FzlA-FtsK interaction occurs between the FzlA C-terminal tail and FtsKC
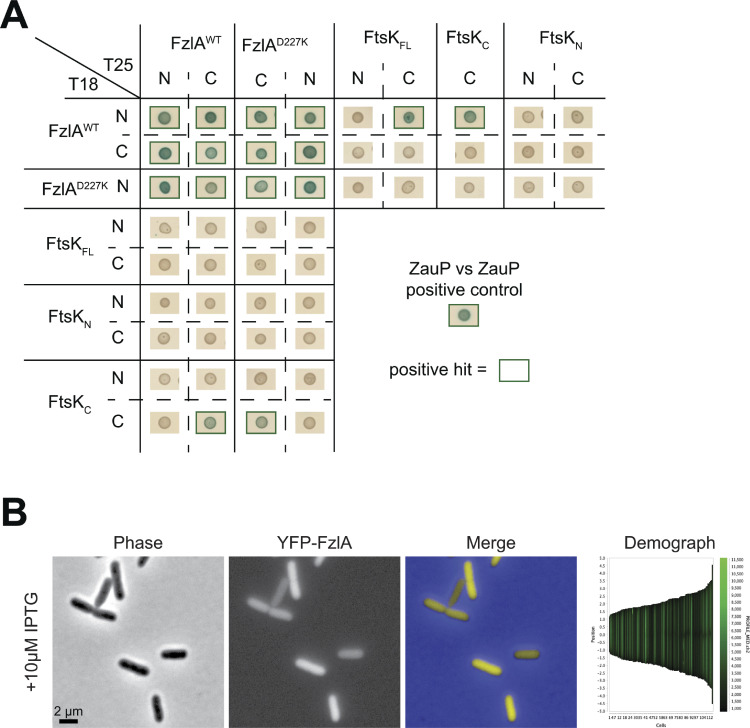
To narrow down the domains responsible for the FzlA-FtsK interaction, we performed bacterial two-hybrid (BTH) analysis in E. coli cells expressing Caulobacter FzlA and FtsK. We first sought to confirm our coIP-MS results for full-length, WT FzlA and FtsK using BTH. FzlA forms a homodimer (Lariviere et al., 2018) and exhibits self-interaction by BTH, serving as a positive control (Fig. 7 C). Indeed, FzlA and full-length FtsK (FtsKFL) were able to interact with BTH (Fig. 7 C). Importantly, a YFP fusion to FzlA does not localize to the division plane when produced in E. coli (Fig. S5 B). This suggests that the observed interaction is not mediated indirectly by recruitment of FzlA and FtsK to the midcell by interactions with E. coli divisome proteins.
Figure S5.
Complete results of BTH analysis. (A) BTH results for interaction between FzlA variants and full-length FtsK or its domains. The adenylyl cyclase subunits T18 (left) and T25 (top) are fused to proteins at the indicated terminus for each row or column. A green box around the representative spot image means that the three biological triplicates were positive for induction of the cAMP-dependent β-galactosidase reporter, indicating a positive interaction. Some data in Fig. 7 C is duplicated in A. (B) Representative phase-contrast, epifluorescence, and merged images, and corresponding demograph of an E. coli strain expressing IPTG-dependent his-yfp-fzlA. The strain was grown to mid-log phase, then induced for 1 h by being grown in the presence of 10 µM IPTG then imaged. Scale bar is 2 µm.
In narrowing down the interacting surfaces on FzlA, we sought to test residues previously determined to be essential for FzlA function. We hypothesized that if the FzlA-FtsK interaction is essential, downstream signaling would likely occur through the essential C-terminal tail (Lariviere et al., 2018). To test this hypothesis, we used a point mutant in the penultimate residue of FzlA (FzlAD227K), a variant that is unable to function in division despite binding FtsZ (Lariviere et al., 2018). Consistent with our hypothesis, FzlAD227K was unable to interact with FtsKFL by BTH, but was still able to self-interact, suggesting the protein is properly folded but that the C-terminus is required to bind FtsK (Fig. 7 C).
FtsKN and FtsKC have independent essential functions (Wang et al., 2006). To determine which domain of FtsK interacts with FzlA, we tested each with FzlA by BTH. The FtsKN construct included the disordered linker up to residue 333, while FtsKC included the remainder of the protein. FzlA was able to interact with FtsKC, but not with FtsKN (Fig. 7 C). To determine if the FzlA C-terminal tail is required for the interaction, we tested the ability of FtsKC and FzlAD227K to interact. FzlAD227K and FtsKC were able to interact by only one orientation, suggesting C-terminal tail mutations disrupt FzlA-FtsK interactions (Fig. 7 C and Fig. S5). Collectively our coIP-MS and BTH approaches indicate that FzlA and FtsK directly interact via the C-terminal tail of FzlA and the DNA translocase domain of FtsK.
FtsK functions upstream of FtsWI
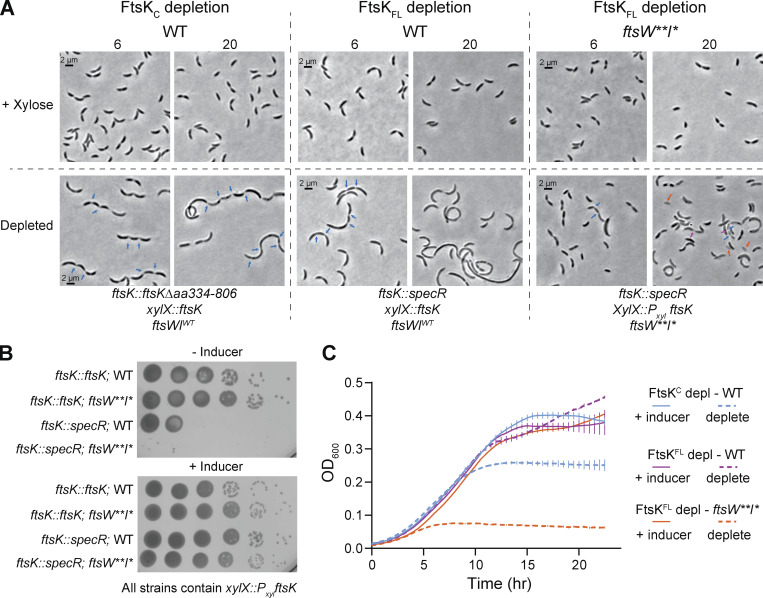
Prior work characterizing Caulobacter FtsK demonstrated that depletion of full-length FtsK or just the C-terminal domain of FtsK resulted in distinct phenotypes. While both were lethal, depletion of full-length FtsK resulted in filamentation while depletion of the C-terminal domain caused cell chaining (Wang et al., 2006). We hypothesized that if FtsK acts downstream of FzlA in an FtsWI activation pathway, in the ftsW**I* background where fzlA is non-essential, FtsWI should also be less dependent on FtsK-mediated activation. While ftsK remains essential in an ftsW**I* background based on comparative transposon sequencing (Lariviere et al., 2019), we reasoned that examining the phenotype associated with depletion of FtsK in an ftsW**I* background could reveal a genetic relationship between ftsK and ftsWI. We generated strains with xylose-dependent expression of ftsK in which ftsKFL was placed at the xylX locus and native ftsK was replaced with a spectinomycin resistance cassette at the native locus in both the WT (EG3332) and ftsW**I* (EG3333) backgrounds. We also examined the phenotype of the previously characterized FtsKC depletion strain (LS4202/EG2996; Wang et al., 2006).
We confirmed the reported depletion phenotypes for FtsKFL and FtsKC, which were predominantly filamentous and chained cells, respectively (Fig. 8 A, chaining: blue arrows). In liquid media, cells depleted of full-length FtsK in a WT background still experienced an increase in absorbance, likely owing to continued growth of filamentous cells. However, FtsK depletion was lethal on solid media (Fig. 8, B and C). Interestingly, cells depleted of FtsKFL in an ftsW**I* background were not filamentous. Instead, they were short, with pleiotropic phenotypes (Fig. 8 A) including extended constrictions (magenta arrows), chaining (blue arrows), and lysis (orange arrows). Despite the presence of cells of normal length, FtsK depletion in the ftsW**I* background was lethal, indicating an essential function of FtsK in addition to its role in regulating FtsWI. Neither FzlA nor FtsK depends on each other or on FtsW for localization to midcell (Fig. S3, B and C), and we previously demonstrated that FtsW localization is independent of FzlA (Goley et al., 2011). These data suggest that the functional interactions among these proteins are not as simple as recruitment to the divisome. We propose that FtsK lies between FzlA and FtsWI in a constriction activation pathway and that input from FtsK may couple chromosome segregation to constriction activation.
Figure 8.
FtsK acts upstream of FtsWI and is required for constriction initiation. (A) Representative phase-contrast images of strains during depletion of FtsKC or FtsKFL in either WT or ftsW**I* backgrounds. Cells were grown in the presence of 0.3% xylose (top row, ftsK induced) or 0.2% glucose (bottom row, FtsK or FtsKC depleted) with images taken at 6 and 20 h after depletion. FtsKC depletion, EG2996. FtsKFL depletion in WT, EG3332. FtsKFL depletion in ftsW**I*, EG3333. (B) Spot dilutions of strains with xylose-inducible ftsK expression in the WT or ftsW**I* background, in the absence or presence of native ftsK. Plates contain either 0.2% glucose (−Inducer) or 0.3% xylose (+Inducer). Row 1, EG3299. Row 2, EG3300. Row 3, EG3332. Row 4, EG3333. (C) Growth curves of strains dependent on xylose for ftsK expression (same as A). Solid lines indicate growth in the presence of 0.3% xylose. Dashed lines indicate growth in the presence of 0.2% glucose. FtsKC depletion, EG2996. FtsKFL depletion in WT, EG3332. FtsKFL depletion in ftsW**I*, EG3333. Points are the mean of three technical replicates at that timepoint, error bars are SEM. Shown is a representative replicate of a biological triplicate.
Proper regulation of FzlA-mediated signaling is critical to prevent DNA damage
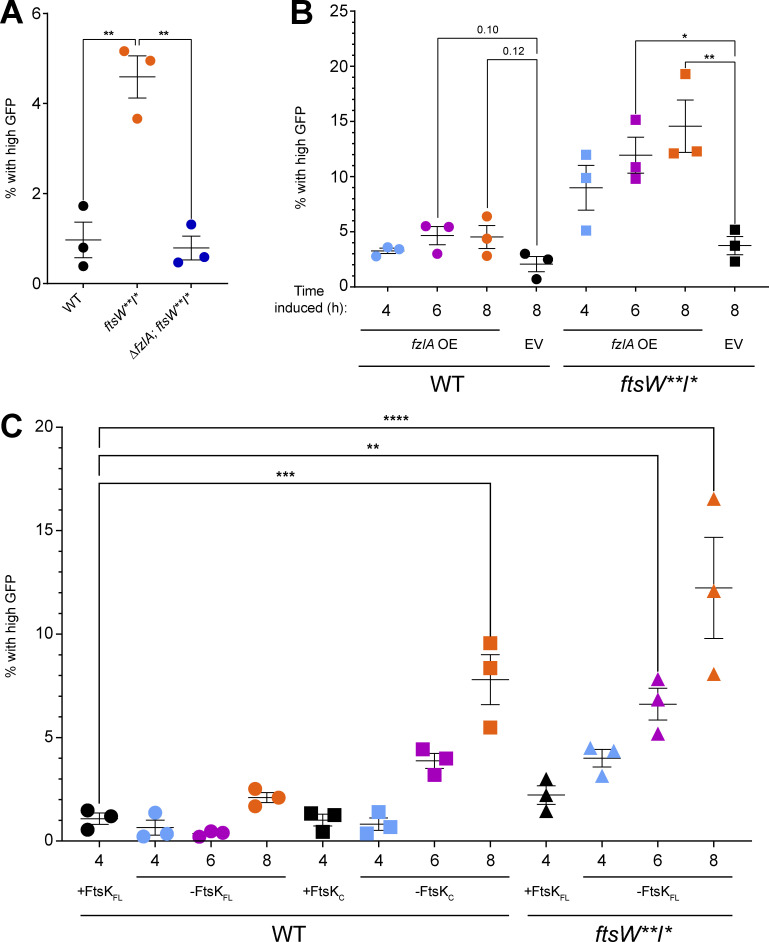
Cell chaining is observed in strains that have cell separation (Meier et al., 2017) or chromosome segregation defects (Wang et al., 2006; Yu et al., 1998). We hypothesized that misregulation of FzlA-FtsK–mediated signaling in a background with hyperactive constriction traps DNA at the division site, preventing the completion of division and causing DNA damage. To test this, we leveraged a previously characterized PsidA-gfp reporter construct (Modell et al., 2014). The sidA promoter is sensitive to DNA damage, leading to production of GFP when DNA damage occurs. To obtain a baseline of DNA damage in Caulobacter, we introduced the reporter construct into a WT background (EG3653). Since DNA damage is rare in a WT background, expression was relatively low. To compare among different strains, we set a threshold for “high GFP” (i.e., DNA damage) as above the 99th percentile of WT GFP values.
First, we introduced the DNA damage reporter construct into ftsW**I* (EG3655) and ∆fzlA, ftsW**I* (EG3667) backgrounds. The hyperconstricting ftsW**I* population had a modest increase in DNA damage (Fig. 9 A and Table S3, 4.59%). Interestingly, the ∆fzlA, ftsW**I* population had WT levels of DNA damage (Table S3 and Fig. 9 A, 0.79%), which suggests that hyperconstriction is the underlying cause of DNA damage, not insensitivity of FtsW**I* to the DNA damage inhibitors SidA and DidA (Modell et al., 2011, 2014).
Figure 9.
Misregulation of the FzlA-FtsK-FtsWI pathway results in DNA damage. (A–C) Percentage of cells with high levels of GFP from the PsidA-gfp reporter. Plots comparing high GFP-producing cells between (A) WT (EG3653), ftsW**I* (EG3655), and ΔfzlA; ftsW**I* (EG3667) backgrounds or (B) strains containing either an EV control or fzlA-overexpression (OE) construct in WT (EV: EG3658, fzlA OE: EG3664) or ftsW**I* (EV: EG3666, fzlA OE: EG3672) backgrounds in the presence of 0.3% xylose for 4, 6, and 8 h. Averages of biological triplicates are shown and were used for statistical analysis. (C) Plot comparing high GFP-producing cells grown in 0.3% xylose for 4 h of a full-length FtsKFL depletion strain in a WT background (EG3904 + xyl 4hr) to various depletion strains grown in the presence or absence of 0.3% xylose (EG3904: FtsK depletion in WT, EG3900: FtsKC depletion in WT, EG3902: FtsK depletion in ftsW**I*) at 4, 6, and 8 h. Averages of biological triplicates are shown and were used for statistical analysis. *P < 0.05. **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine if hyperconstriction due to FzlA overproduction also leads to DNA damage, we introduced the reporter into our fzlA-overexpressing and EV strains in WT and ftsW**I* backgrounds. The EV control in a WT (EG3658) background had WT levels of DNA damage (Fig. 9 B and Table S3, 2.1%). The FzlA-overproducing WT strain (EG3664) experienced a modest increase in DNA damage at 4.6% at 6 and 8 h of induction (Fig. 9 B and Table S3). We next assessed DNA damage in cells overproducing FzlA (EG3672) compared with the EV control (EG3666) in the ftsW**I* background. Considering FzlA overproduction in this background led to cell chaining, we hypothesized the DNA damage may be greater than in WT. In line with this hypothesis, FzlA overproduction in the ftsW**I* background significantly increased DNA damage when compared with the EV control in a time-dependent manner (Fig. 9 B and Table S3, 4.6% at 4 h, 11.94% at 6 h, 14.56% at 8 h). We conclude that hyperactivation of constriction by FzlA overproduction leads to DNA damage.
We next tested whether modulation of FtsK causes DNA damage by introducing the DNA damage reporter into the xylose-dependent FtsKFL depletion strain (EG3904; Fig. 8). We set a threshold for high GFP as above the 99th percentile of GFP values observed in this strain grown in the presence of xylose to induce ftsKFL. We then introduced the DNA damage reporter into the FtsKC (EG3900) depletion strain in a WT background. We hypothesized that depleting FtsKFL in a WT background would not result in DNA damage since these cells fail to initiate constriction. Supporting our hypothesis, we observed no change in DNA damage during depletion (Fig. 9 C and Table S3, 0.647% at 4 h, 0.360% at 6 h, 2.10% at 8 h) when compared to the replete condition (Fig. 9 C and Table S3, 1.08%). We next examined the effect of depleting FtsKC as this results in cell chaining. Indeed, there were higher levels of DNA damage at 6 (Fig. 9 C and Table S3, 3.88%) and 8 h (7.80%) of depletion compared with the replete control (1.08%), with DNA damage increasing over time as FtsKC was depleted.
Finally, we tested the effect of depleting FtsKFL in an ftsW**I* background since these cells experience chaining and extended constrictions. We introduced the DNA damage reporter into an ftsW**I* strain with the only copy of ftsKFL under the control of the xylose-dependent promoter (EG3902). When we depleted FtsK in this background, we observed a time-dependent increase in DNA damage (Fig. 9 C and Table S3, 4.00% at 4 h, 6.62% at 6 h, 12.2% at 8 h) compared with the control (Fig. 9 C and Table S3, 1.08%). Collectively, these data indicate that misregulating the FzlA-FtsK-FtsWI pathway results in DNA damage due to improper coupling of chromosome segregation to constriction.
Discussion
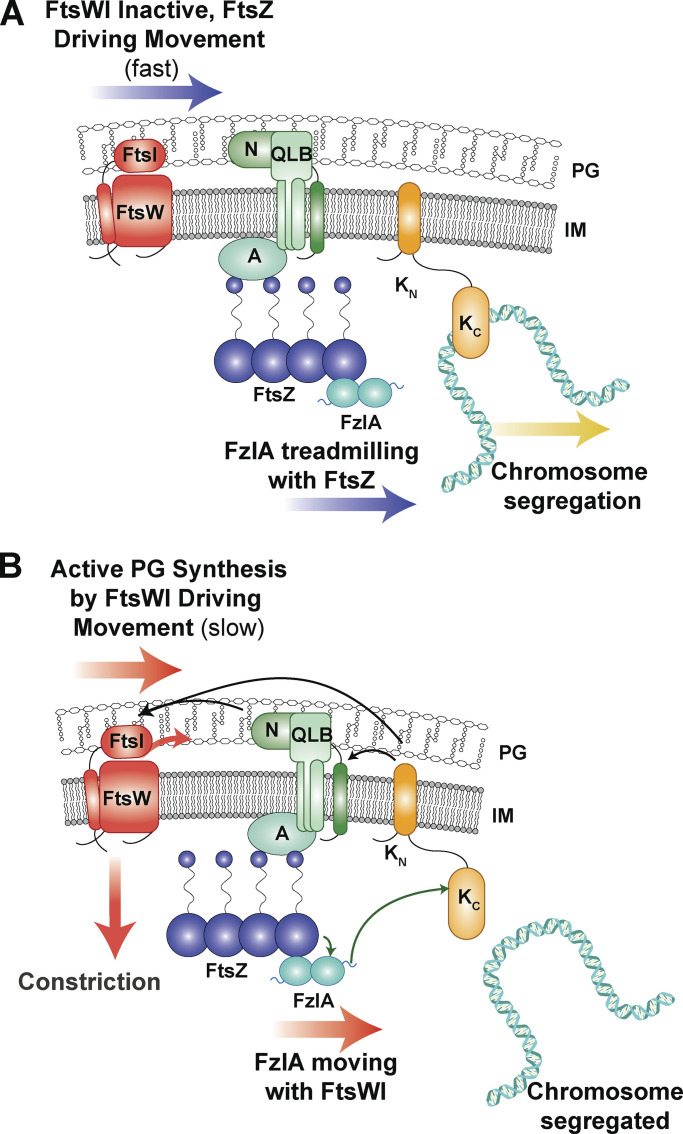
The specifics of bacterial constriction activation can vary from species to species, but the functional requirements to select the division plane, dynamically position the divisome, segregate the chromosomes, and activate constriction are essential and conserved. Our work describes a key role for FzlA in signaling to activate FtsWI in coordination with chromosome segregation. FzlA activates constriction during division by signaling to convert FtsW molecules into an active, slow-moving state (Figs. 1, 2, 3, 4, and 5). The FzlA activation pathway signals through FtsK, an essential protein that segregates sister chromosomes during division (Figs. 7 and 8). When this signaling is misregulated through hyperactivating mutations or overproduction of FzlA, the cell suffers cell envelope defects (Lariviere et al., 2019; Modell et al., 2014; Fig. 6) and DNA damage (Fig. 9), likely due to defects in coordinating chromosome segregation with division. We propose that, prior to activation, inactive FtsWI is either stationary or fast moving through indirect association with treadmilling FtsZ clusters (Fig. 10 A). FzlA binds FtsZ and single FzlA molecules may infrequently move quickly by diffusion or by dynamic association with FtsZ filament ends or, more frequently, remain stationary (but treadmilling) by association with the interior of FtsZ filaments. We propose that when FtsK is not bound to DNA (Fig. 10 B), FzlA and FtsK can interact via FtsKC and the C-terminal tail of FzlA. The FzlA-FtsK interaction signals to license FtsWI activation, likely through additional factors. Ultimately, FtsWI is converted to an active state that moves slowly, driven by PG synthesis (Fig. 10 B). FzlA can move together with the activated FtsWI complex.
Figure 10.
FzlA binds to FtsZ and signals through FtsK to shift FtsW to a slow-moving, active state. (A) When FtsK is actively segregating chromosomal termini from the division plane, it has limited interaction with FzlA and signaling downstream to activate FtsWI does not occur. Inactive FtsW molecules either move with FtsZ clusters as they treadmill about the division plane or are stationary. FzlA molecules treadmill with FtsZ or remain stationary. (B) When FtsK is not bound to DNA, the FzlA C-terminal tail can bind to FtsKC to signal downstream for FtsWI activation, thereby coupling chromosome segregation to constriction progression. (A and B) Black arrows indicate hypothesized signaling. Green arrows indicate direct interactions confirmed in Caulobacter. Other divisome components depicted for hypothesized downstream interactions: A, FtsA; QLB, FtsQLB complex; N, FtsN.
Though we were initially motivated to understand how FzlA regulates constriction, our study led us to first define the dynamics of FtsW in Caulobacter. The question of how the dynamics of PG synthases are linked to their activity and FtsZ treadmilling has been intensely probed across a number of bacterial species in recent years. Of those, the only Gram-negative organism studied was E. coli, in which the two-track model for PG synthase movement was proposed (Yang et al., 2021). This is distinct from S. aureus and S. pneumoniae in which PG synthase movement is uncoupled from FtsZ treadmilling (Perez et al., 2019; Schäper et al., 2023, Preprint) or B. subtilis, where coupling is reported to varying extents depending on the experimental conditions (Bisson-Filho et al., 2017; Whitley et al., 2023, Preprint). Our work indicates that, like E. coli, Caulobacter FtsW moves in two populations: slow, active molecules driven by PG synthesis, and fast, inactive molecules driven by FtsZ treadmilling. For both FzlA and FtsW, we observed transitions from the fast- or slow-moving populations to the stationary state and vice versa. Though it is difficult to quantify the frequency of these events, this suggests the PG synthase complex can transfer between the FtsZ and PG track. Given the evolutionary distance between E. coli (a ɣ-proteobacterium) and Caulobacter (an α-proteobacterium), it may be that the two-track model is a widespread mode of divisome dynamics among proteobacteria.
Our findings also provide a deeper understanding of the mechanisms of FtsWI hyperactivation and its consequences. We observed that active FtsW** synthesized PG for longer periods than FtsW. Conversely, FzlA did not impact the processivity of active FtsW or FtsW**, suggesting increased processivity results from a mechanism distinct from that of FzlA-dependent activation. The mutations of FtsW** may perturb association with a regulator that modulates processivity, for example. An obvious candidate is the FtsQLB complex, based on the observations made in E. coli and P. aeruginosa (Tsang and Bernhardt, 2015; Marmont and Bernhardt, 2020; Park et al., 2020), though further experiments are required to test this hypothesis. Alternatively, mutation of FtsWI may directly impact processivity of the enzyme.
We discovered that FzlA signals to convert FtsW to an active, slow-moving state and that upstream signaling from FzlA is limiting for FtsW activation (Fig. 10 B). Our observations for FzlA are, in some ways, similar to observations made for FtsN in hyperactive ftsB mutant (ftsBE56A) and WT backgrounds in E. coli (Lyu et al., 2022). Expression of ftsBE56A hyperactivates the PG synthases, causes cell shortening, and renders ftsN non-essential. The double mutant of ΔftsN, ftsBE56A results in cell lengths more similar to WT E. coli, and the PG synthases are active less often than in the ftsBE56A background with FtsN. FtsN depletion in an otherwise WT background also results in cell filamentation, and the PG synthases are more often inactive, consistent with FtsN being an FtsW activator. These results mirror many of our FzlA results and may be a common feature of factors that signal to activate FtsWI. However, there are important differences between FzlA and FtsN and we propose that they act at distinct stages of divisome activation in α-proteobacteria to couple division to other cellular processes. FtsN is a transmembrane protein with a PG-sensing SPOR domain that is proposed to couple PG synthesis to amidase activity at the division site (Yahashiri et al., 2015; Gerding et al., 2009). Based on genetics in E. coli, FtsN is thought to bind directly to the FtsQLB complex, which directly interacts with and activates FtsWI (Park et al., 2021; Gerding et al., 2009; Marmont and Bernhardt, 2020; Käshammer et al., 2023; Britton et al., 2023). Conversely, FzlA is a soluble FtsZ-binding protein that also binds the chromosome-segregation factor FtsK. Considering that Caulobacter has essential homologs of FtsN and FtsQLB, we propose that FzlA and FtsK provide an additional layer of regulation that serves to integrate information from the Z-ring and chromosome with constriction.
In Caulobacter, the essential divisome components include FtsZ, FtsA, FzlA, FtsK, FtsN, FtsQLB, and FtsWI (Goley et al., 2011). FzlA is restricted to the α-proteobacteria and FtsN is restricted to proteobacteria, but the remainder of these proteins are widely conserved across bacterial phyla. Our work characterizing the FzlA-FtsK interaction identifies, for the first time, where FtsK integrates in the constriction activation pathway in α-proteobacteria. The FzlA-FtsK interaction is likely conserved throughout α-proteobacteria, as residues 223, 227, and 228 of the FzlA C-terminal tail are invariant in FzlA homologs (Lariviere et al., 2018, 2019). FzlA homologs are even found in the Rickettsiales, which have streamlined genomes and are often outliers among the α-proteobacteria in conservation of factors that are essential in Caulobacter.
Outside of α-proteobacteria, FtsK and the family of FtsK/SpoIIIE/HerA homologs are widespread and often essential, even extending to archaea (Bigot et al., 2007). In E. coli, only FtsKN is essential, which suggests that its role in the constriction activation pathway is conserved, but that redundant chromosome maintenance mechanisms support growth in the absence of FtsKC (Draper et al., 1998). Indeed, E. coli encodes a nucleoid occlusion system as an additional chromosome maintenance mechanism. SlmA binds to the nucleoid and prevents Z-ring assembly locally, which prevents the cell from constricting over the nucleoid (Bernhardt and de Boer, 2005). Caulobacter and most other α-proteobacteria lack nucleoid occlusion systems. In addition, in E. coli the MatP-ZapB-ZapA protein network that connects the FtsZ ring to the chromosome is proposed to act as a “brake” to prevent premature cell wall constriction before chromosome segregation (Buss et al., 2015). Caulobacter encodes an analogous system composed of ZapT-ZauP-ZapA that links the chromosomal termini to the Z-ring (Woldemeskel et al., 2017; Ozaki et al., 2020). However, in Caulobacter these proteins help to condense and position the Z-ring and do not appear to regulate constriction rate. We therefore propose that FzlA functions in α-proteobacteria as the primary mechanism to couple constriction to chromosome segregation through FtsK.
In Caulobacter, the FtsZ-FzlA-FtsK-FtsWI pathway coordinates constriction with chromosome dimer resolution and segregation of termini. We propose that FzlA binds FtsK and signals to activate FtsWI when FtsK is not actively engaged with DNA. The likelihood of this interaction—and therefore of signaling to activate PG synthesis—would increase as the bulk of the chromosome is cleared from the division plane. In this way, FzlA-FtsK would act as a “pacer” of constriction. FzlA binding to FtsZ is also required for constriction, but it is unclear if this interaction serves simply to position FzlA at the division site or plays a more active regulatory role. The precise molecular choreography dictating interactions among and dynamics of the components of our proposed FzlA-dependent pathway of constriction regulation are exciting avenues for future work.
Materials and methods
Caulobacter and E. coli growth media and conditions
C. crescentus NA1000 cells were grown at 30°C in peptone-yeast extract (PYE) media. E. coli NEB Turbo (Catalog #C2986K; NEB), BTH101, TOP10, and Rosetta cells were grown at 37°C in Luria-Bertani (LB) medium. Xylose was used at final concentrations of either 0.001% or 0.3% (wt/vol) as indicated. Glucose was used at final concentrations of either 0.001% or 0.2% (wt/vol) as indicated. For depletion strains, cells were grown in PYE supplemented with xylose as indicated prior to being washed with plain PYE medium three times and resuspended in the PYE medium supplemented with the stated concentrations of glucose or xylose. Solid media included 1.5% (wt/vol) agar. Antibiotics were used in liquid (solid) medium at the following concentrations for Caulobacter: kanamycin, 5 (25) µg/ml; gentamycin, 1 (5) µg/ml; spectinomycin, 25 (100) µg/ml; and oxytetracycline, 1 (2) µg/ml. Streptomycin was used at 5 µg/ml in solid media. E. coli antibiotics were used in liquid (solid) medium as follows: kanamycin, 30 (50) µg/ml; gentamycin 15 (20) µg/ml; spectinomycin, 50 (50) µg/ml; ampicillin, 50 (100) µg/ml; and oxytetracycline, 12 (12) µg/ml. For growth curves, a Tecan Infinite M200 Pro plate reader measured absorbance every 30 min at OD600 of a 100 μl culture, at a starting OD600 of 0.05, in a 96-well plate in biological and technical triplicate with intermittent shaking. For spot dilution assays, mid-log cells were diluted to an OD600 of 0.05 and serially diluted up to 10−5 before spotting 5 μl onto a PYE plate with indicated inducer and/or antibiotic. Plates were incubated at 30°C for 48 h. For time-lapse microscopy or SMT experiments of FzlA-overproducing cells, induction with 0.3% xylose occurred for 1 h before synchrony and throughout the experiment.
Synchrony
Synchrony was performed as previously described (Goley et al., 2011). Cells were grown overnight in 15 ml of PYE medium to an OD600 between 0.1 and 0.5, harvested by pelleting at 6,000 rpm at 4°C, resuspended in 750 μl of ice-cold M2 salts, and combined with 750 μl of Percoll (Catalog #45-001-748; Thermo Fisher Scientific). The cell suspension was centrifuged at 20,000 × g for 20 min at 4°C. The swarmer band was isolated, transferred to a fresh tube, and pelleted at 8,000 × g for 2 min. The swarmer pellet was washed twice with 1 ml of ice-cold M2 salts and pelleted each time at 8,000 × g for 2 min. The swarmer cells were resuspended in PYE with appropriate additives and grown at 25°C.
Phase-contrast and standard epifluorescence microscopy
Cells in exponential phase of growth were spotted on pads made of 1% agarose resuspended in water or PYE medium, supplemented with 0.3% xylose when appropriate, and imaged using a Nikon Eclipse Ti inverted microscope (RRID:SCR_021242) equipped with a Nikon Plan Fluor 100X (NA1.30) oil Ph3 objective and Photometrics CoolSNAP HQ2 cooled CCD camera. Time-lapse imaging was performed as previously described (Lariviere et al., 2019), using Gene Frames (Catalog #AB0577; Thermo Fisher Scientific) to ensure a tight seal during imaging. After synchrony, cells were resuspended in PYE with the appropriate additives, and a stopwatch was started to begin measuring preconstriction time. The cell suspension was spotted on agarose pads. After the pad was allowed to dry sufficiently, the top coverslip was adhered to the gene frame and time-lapse imaging was initiated. Images were acquired every 5 min for 4 h at room temperature using NIS Elements software (RRID:SCR_014329) and processed using FIJI (RRID:SCR_002285) with the MicrobeJ (Ducret et al., 2016) plugin or using Adobe Photoshop (RRID:SCR_014199).
Time-lapse microscopy analysis for growth metrics comparison
To determine dimensions of log-phase cells, cell length and width were measured using FIJI (RRID:SCR_002285; Schindelin et al., 2012) and MicrobeJ (Ducret et al., 2016) software, similar to as previously described (Lariviere et al., 2018). Constriction rate and elongation rate were also determined using MicrobeJ. MicrobeJ software allowed for tracking of cells imaged by time-lapse microscopy throughout the division process, with automatic detection of constriction initiation and manual determination of cell separation. Cell length was found for cells at each time point, cell width was found at the site of constriction, and constriction time was calculated by multiplying the number of frames in which constriction was detected by 5 (since images were acquired every 5 min), allowing for calculation of constriction and elongation rates. Lethal division events were quantified manually. Prism was used for graphing and statistical analysis of calculated terms. Statistical analysis for Superplots involved one-tailed Mann–Whitney tests, based on the hypothesis that FzlA overproduction would enhance constriction. All cells that were in focus were included in these analyses.
Immunoblot analysis
Immunoblot analysis was performed using standard procedures with a 1:6,666 dilution of affinity-purified α-FzlA antibodies (Goley et al., 2010), a 1:2,500 dilution of α-MreB antisera (Beaufay et al., 2015), a 1:2,500 dilution of α-RFP antisera (Chen et al., 2005), a 1:1,000 dilution of α-FLAG M2 antibodies (Catalog #F3165-1MG; Millipore Sigma), and/or 1:10,000 dilution of HRP-labeled α-rabbit secondary antibodies (Catalog #170-6515; BioRAD) on nitrocellulose membranes. ClarityWestern Electrochemiluminescent substrate (Catalog #170-5060; BioRAD) was for visualization via an Amersham Imager 600 RGB gel and membrane imager (GE).
Advanced epifluorescence imaging and SMT of Halo-FtsW and Halo-FzlA
Except for the FzlA depletion experiment, all SMT experiments were performed after synchrony. For experiments comparing FtsW movement in cells producing FtsZWT versus FtsZD216A, cells were washed and grown without inducer for 1 h, synchronized, then grown in PYE with vanillate or xylose to induce ftsZWT or ftsZD216A, respectively, for 1 h before imaging. After synchrony, cells were labeled with Janelia Fluor 646 (a gift from Dr. Luke Lavis, Howard Hughes Medical Institute, Chevy Chase, MD, USA; Catalog #GA1120; Promega) at an empirically determined concentration for each sample to obtain single-molecule level labeling, ranging from 0.2 to 10 nM in PYE with additives as required (xylose or glucose). The cell suspension was incubated in the dark while shaking at 25°C for 15 min before washing away the dye with three washes of 1 ml PYE. Cells were resuspended in fresh PYE and allowed to continue growing until the time of the experiment. To inhibit cell wall synthesis, Fos was added to synchronized cells at 100 µg/ml for 30 min before the images were acquired. Under all conditions, cells were imaged no later than 90 min after synchrony, with the imaging beginning 1 h after synchrony.
SMT experiments were performed as previously described (Yang et al., 2021). Briefly, single JF646-labeled Halo-FtsW or Halo-FzlA molecules were detected on an Olympus IX-81 inverted microscope equipped with a UPlanApo 100× TIRF (1.50NA/oil) objective and an Andor iXon 897 Ultra EM-CCD camera in epifluorescence-illumination mode using a Coherent 647-nm laser at ∼2 W•cm−2 power density. Brightfield and fluorescent images were acquired using Metamorph 7.8.13.0 software (RRID:SCR_002368) at room temperature. The camera’s electron-multiplying gain was turned to 300 with a preamplifier gain setting of 3 and a digitizer set to 16-bit. Baseline clamp was activated, with a baseline offset set to 100. The focal plane was placed in the middle of the cell to capture the most moving molecules at the division plane. SMT images were taken continuously with 500-ms exposure time for 200 s.
Fluorescent spots of single FtsW or FzlA molecules were first localized using two-dimensional Gaussian fitting in an ImageJ (1.52p; RRID:SCR_003070; Schneider et al., 2012) plug-in, ThunderSTORM (dev-2016-09-10-b1; Ovesný et al., 2014). A bandpass filter (80–250 nm) for Sigma-Aldrich was applied to remove the single-pixel noise (small Sigma-Aldrich) and out-of-focus (big Sigma-Aldrich) molecules. To focus on the FtsW or FzlA during cell division, we only analyzed the molecules located at midcell or places with observable invagination. In the case of the FzlA depletion strain (EG3523) without obvious constrictions, ZapA-mNG–marked Z-rings were used to identify divisome-associated FtsW trajectories. Those localizations were then linked to trajectories using a home-built Matlab 2020a (RRID:SCR_001622) script (Yang et al., 2023) adopting the nearest neighbor algorithm as previously described (Sbalzarini and Koumoutsakos, 2005; Yang et al., 2021). For the movement of each molecule, the rate for each segment was measured and recorded, but for calculating the proportion of moving molecules, the molecule was counted once. The localization error of our microscope setup was ∼23 nm based on fitting the nearest-neighbor distance between localizations of stationary molecules (Endesfelder et al., 2014).
The unwrapped and corrected trajectories described above were then segmented to determine whether Halo-FtsW or Halo-FzlA molecules in a segment are stationary or moving directly as previously described (Yang et al., 2021). The cumulative probability density function (CDF) of the directional moving speeds was further calculated and fit to a two-population model (of log-normal distribution) as described previously (Yang et al., 2021):
where the v is the FtsW (or FzlA) moving speed and P1 is the percentage of the first population. For a single-population model, P1 = 1. The two parameters μ and σ are the natural logarithmic mean and standard deviation of the speed. erf is the error function. To determine the FtsW fast-moving speed coupled with FtsZ’s treadmilling speed, we first fit the CDF of the Fos-treated condition (Fig. 2, C iii and iv) without fixing any parameters. The median speed of the fast-moving population (µ2) was considered as the FtsZwt treadmilling speed and fixed in other conditions. The width of the fast-moving speed (σ2) distribution was not fixed. The residuals (such as Fig. 2, C ii and iv) were calculated by subtracting the experimental CDF from the fit curve. For FzlA, two-population fitting was performed as above, except fitting did not fix any parameters other than setting a minimum cutoff of 18 nm/s for the fast population.
To visualize the relationship between the speed and lifetime of active (slow-moving) molecules (Fig. 3 C; Fig. 4, B and D; and Fig. 5 C), we established a threshold of 20 nm/s as the intersection point of the two fitted curves representing the fast (blue) and slow (orange) populations (Fig. 2 B). Molecules with a speed <20 nm/s are more likely to be active.
Statistical tests
To substantiate the performance of both single- and double-population fitting approaches, we utilized the Akaike Information Criterion (AIC) to assess the quality of each fit. Assuming that the fitting error follows a normal distribution, we initially computed the log-likelihood of each fitting based on the residuals using the formula:
Here, L represents the likelihood of the fitting, n denotes the number of data points, S signifies the standard deviation of the residuals, and resn represents the sum of the squared residuals.
In this formula, Np corresponds to the number of parameters specific to the chosen model. For the single-population fitting, Np equals 3, while for the two-population fitting, Np equals 5.
CoIP
CoIP was performed as previously described (Bowman et al., 2008; Lariviere et al., 2018). All coIP steps, unless designated, were performed at 4°C. CoIP buffer consists of 20 mM HEPES pH 7.5, 100 mM NaCl, and 20% glycerol and one Pierce Protease Inhibitor Mini Tablet, EDTA Free, (A32955; Thermo Fisher Scientific) per liter. Cells were harvested during exponential growth by centrifugation at 6,000 × g at 4°C. Cells were washed with coIP buffer, pelleted at 6,000 × g, and resuspended in coIP buffer. Dithiobis(succinimidyl propionate) (DSP) was used to crosslink free amines. DSP was resuspended at 10 mg/ml in DMSO and added to the cell suspension to reach a final concentration of 2 mM of DSP in 0.08% DMSO for 1 h at 4°C and 10 min at room temperature. The reaction was quenched by adding 1 M Tris-HCl pH 7.5 to a final concentration of 20 mM for 10 min. After quenching, cells were lysed by three passes of an EmulsiFlex-C3 at 16,000 psi. Cell membranes were solubilized by adding sodium deoxycholate to 0.5% wt/vol, Igepal CA-630 to 1% wt/vol, and EDTA to a 2 mM final concentration. Insoluble cellular debris was removed by pelleting at 4,500 × g for 10 min. For each sample, up to 40 ml of supernatant of each sample was transferred to a fresh tube and normalized based on OD to the lowest density sample. To each tube, 100 μl of washed α-FLAG resin (Catalog #A2220-5ML; Millipore Sigma) was added before incubating on a rotator overnight. Afterward, the resin was pelleted at 6,000 × g and washed to remove non-specific bound proteins. Proteins were eluted by incubating the resin with 100 μl of free FLAG peptide (Catalog #F3290-4MG; Millipore Sigma) at a concentration of 0.5 mg/ml for 15 min, three times. The pooled elution was either provided to the Mass Spectrometry Core Facility at Johns Hopkins University or prepared for western blot analysis.
BTH
Plasmids containing fusions to T18 and T25 were cotransformed into BTH101 competent cells and plated onto LB agar supplemented with ampicillin (100 μg/μl) and kanamycin (50 μg/μl) and grown overnight at 30°C. Biological triplicates of each cotransformation were grown overnight at 30°C in LB including with ampicillin (100 μg/μl), kanamycin (50 μg/μl), and IPTG (0.5 mM). After overnight incubation, 2 μl of each was spotted on LB agar plates supplemented with ampicillin (100 µg/μl), kanamycin (50 µg/μl), X-gal (40 μg/ml), and IPTG (0.5 mM) and incubated for 2 d at 30°C. Blue colonies were considered positive protein-protein interactions.
DNA damage quantification
To quantify DNA damage using the PsidA-egfp reporter, average fluorescence intensity for each cell in three biological replicate experiments per strain was measured using MicrobeJ. We calculated high GFP intensity, which we considered above the 99th percentile of the specified control strain GFP values, by first sorting the average fluorescent intensities for each replicate from lowest to highest. For the fzlA overexpression or deletion and ftsW**I* strains, we compared fluorescent intensity values to the 99th percentile of WT GFP values. For the FtsK depletion strains, we compared fluorescent intensity values to the 99th percentile of the replete condition of the FtsKFL depletion strain in a WT background grown for 4 h in the presence of 0.3% xylose. We then multiplied the total number of cells analyzed for each control replicate by 0.99. The average intensity of that resulting cell number was then recorded. After obtaining the fluorescence intensity of the 99th percentile of each control replicate, we averaged the fluorescence intensity of the three replicates. For all of the strains analyzed, we looked for fluorescence intensity values above the calculated 99th percentile of the control strain average. The number of cells that had higher values was counted and then divided by the total number of cells analyzed for each replicate of each strain. The average percentage of cells above the 99th percentile of the control strain fluorescent intensity was then calculated by averaging the percentages of each replicate for each strain.
Online supplemental material
Fig. S1 contains western blots that confirm the protein levels in various strains used throughout this study. Fig. S2 demonstrates example trajectories and speed measurements generated by SMT of Halo-FtsW. Fig. S3 illustrates that ZapA-mNG is produced and localized to the division plane, validating its use for locating Z-rings in FzlA-depleted cells. Fig. S3 also demonstrates the independence of FzlA and FtsK localization to the division plane from each other and from FtsW. Fig S4 is population fitting of the SMT data from Fig. 4. Fig. S5 displays the tested combinations of FzlA and FtsK variants in each orientation for interaction by BTH and shows YFP-FzlA does not localize to the site of division in E. coli cells. Table S1 includes the SMT results of Halo-FtsW and Halo-FzlA in the various backgrounds and conditions referred to throughout the text. Table S2 presents the results from MS-based protein identification of eluent from the 3xFLAG-FzlA and WT coIP samples. Table S3 provides the DNA damage results from the various strains and conditions referred to in Fig. 9. Table S4 shows strains and plasmids used in this study. Video 1 is an example time-lapse of a Caulobacter cell constricting and dividing. Videos 2, 3, and 4 are examples of single molecules of FtsW** (Video 2) and FtsW (Videos 3 and 4) that illustrate example movements by SMT. Videos 5, 6, and 7 are examples of the three constriction defects (Video 5, both daughters halt growth after division; Video 6, one daughter halts growth after division; Video 7, one daughter lyses after division) observed during FzlA overproduction.
Supplementary Material
shows the summary of SMT of Halo-FtsW and Halo-FzlA variants with varying states of activation.
shows proteins identified by MS that are fivefold enriched in relative abundance in 3xFLAG-FzlA eluate compared to WT.
shows the summary of DNA damage reporter experiments.
shows strains and plasmids used in this study.
is the source file for Fig. 7.
is the source file for Fig. S1.
Acknowledgments
The authors would like to thank Lucy Shapiro (Stanford University, Stanford, CA, USA), Michael Laub (Massachusetts Institute of Technology, Cambridge, MA, USA), Régis Hallez (University of Namur, Namur, Belgium), and Patrick Viollier (University of Geneva, Geneva, Switzerland) for strains and antibodies, and Luke Lavis (Janelia Research Campus, Ashburn, VA, USA) for Janelia Fluor dyes. We thank members of the Goley and Xiao laboratories (Johns Hopkins University, Baltimore, MD, USA) for helpful discussions. We thank Di Yan of the Yang lab (University of Science and Technology of China, Hefei, China) for polishing figures.
This study was supported by the National Institutes of Health under awards R35GM136221 (E.D. Goley), R01GM108640 (E.D. Goley), R35GM136436 (J. Xiao), and T32GM144272 (training grant support of I.P. Payne) and by the National Natural Science Foundation of China (award 32270035 to X. Yang) and Anhui Provincial Natural Science Foundation (award 2208085MC40 to X. Yang).
Author contributions: C.R. Mahone: Conceptualization, Formal Analysis, Investigation, Methodology, Resources, Visualization, Writing—Original Draft, and Review & Editing; I.P. Payne: Formal Analysis, Investigation, Methodology, Resources, Visualization, Writing—Original Draft, and Review & Editing. Z. Lyu: Formal Analysis, Investigation, Visualization, and Editing—Review & Editing. J.W. McCausland: Formal Analysis, Investigation, Visualization, and Writing—Review & Editing. J.M. Barrows: Resources, Writing—Review & Editing. J. Xiao: Conceptualization, Funding Acquisition, Project Administration, Supervision, and Writing—Review & Editing. X. Yang: Conceptualization, Formal Analysis, Funding Acquisition, Investigation, Methodology, Visualization, and Writing—Original Draft, Review & Editing. E.D. Goley: Conceptualization, Formal Analysis, Funding Acquisition, Methodology, Project Administration, Resources, Supervision, Visualization, and Writing—Original Draft, Review & Editing.
Data availability
Data are available in the article itself and its supplementary materials. Strains and plasmids generated in this study are available from the corresponding author upon request.
References
- Barrows, J.M., Sundararajan K., Bhargava A., and Goley E.D.. 2020. FtsA regulates z-ring morphology and cell wall metabolism in an FtsZ C-terminal linker-dependent manner in Caulobacter crescentus. J. Bacteriol. 202:e00693-19. 10.1128/JB.00693-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows, J.M., and Goley E.D.. 2021. FtsZ dynamics in bacterial division: What, how, and why? Curr. Opin. Cell Biol. 68:163–172. 10.1016/j.ceb.2020.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows, J.M., Andereson A.S., Talavera-Figueroa B.K., and Goley E.D.. 2023. Intrinsic and extrinsic factors regulate FtsZ function in Caulobacter crescentus. bioRxiv. 10.1101/2023.09.08.556907 (Preprint posted September 08, 2023). [DOI] [Google Scholar]
- Battesti, A., and Bouveret E.. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods. 58:325–334. 10.1016/j.ymeth.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Beaufay, F., Coppine J., Mayard A., Laloux G., De Bolle X., and Hallez R.. 2015. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J. 34:1786–1800. 10.15252/embj.201490730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt, T.G., and de Boer P.A.J.. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52:1255–1269. 10.1111/j.1365-2958.2004.04063.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt, T.G., and de Boer P.A.J.. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell. 18:555–564. 10.1016/j.molcel.2005.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot, S., Sivanathan V., Possoz C., Barre F.X., and Cornet F.. 2007. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 64:1434–1441. 10.1111/j.1365-2958.2007.05755.x [DOI] [PubMed] [Google Scholar]
- Bisson-Filho, A.W., Hsu Y.-P., Squyres G.R., Kuru E., Wu F., Jukes C., Sun Y., Dekker C., Holden S., VanNieuwenhze M.S., et al. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 355:739–743. 10.1126/science.aak9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, G.R., Comolli L.R., Zhu J., Eckart M., Koenig M., Downing K.H., Moerner W.E., Earnest T., and Shapiro L.. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 134:945–955. 10.1016/j.cell.2008.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton, B.M., Yovanno R.A., Costa S.F., McCausland J., Lau A.Y., Xiao J., and Hensel Z.. 2023. Conformational changes in the essential E. coli septal cell wall synthesis complex suggest an activation mechanism. Nat. Commun. 14:4585. 10.1038/s41467-023-39921-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss, J., Coltharp C., Shtengel G., Yang X., Hess H., and Xiao J.. 2015. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 11:e1005128. 10.1371/journal.pgen.1005128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.C., Viollier P.H., and Shapiro L.. 2005. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 55:1085–1103. 10.1111/j.1365-2958.2004.04443.x [DOI] [PubMed] [Google Scholar]
- Coltharp, C., and Xiao J.. 2017. Beyond force generation: Why is a dynamic ring of FtsZ polymers essential for bacterial cytokinesis? BioEssays. 39:1–11. 10.1002/bies.201600179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitch, A.K., and Goley E.D.. 2020. Uncovering unappreciated activities and niche functions of bacterial cell wall enzymes. Curr. Biol. 30:R1170–R1175. 10.1016/j.cub.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewachter, L., Verstraeten N., Fauvart M., and Michiels J.. 2018. An integrative view of cell cycle control in Escherichia coli. FEMS Microbiol. Rev. 42:116–136. 10.1093/femsre/fuy005 [DOI] [PubMed] [Google Scholar]
- Draper, G.C., McLennan N., Begg K., Masters M., and Donachie W.D.. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180:4621–4627. 10.1128/JB.180.17.4621-4627.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, S., Pichoff S., and Lutkenhaus J.. 2016. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl. Acad. Sci. USA. 113:E5052–E5061. 10.1073/pnas.1606656113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret, A., Quardokus E.M., and Brun Y.V.. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1:16077. 10.1038/nmicrobiol.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endesfelder, U., Malkusch S., Fricke F., and Heilemann M.. 2014. A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem. Cell Biol. 141:629–638. 10.1007/s00418-014-1192-3 [DOI] [PubMed] [Google Scholar]
- Evinger, M., and Agabian N.. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294–301. 10.1128/jb.132.1.294-301.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding, M.A., Liu B., Bendezú F.O., Hale C.A., Bernhardt T.G., and de Boer P.A.J.. 2009. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191:7383–7401. 10.1128/JB.00811-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley, E.D., Dye N.A., Werner J.N., Gitai Z., and Shapiro L.. 2010. Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell. 39:975–987. 10.1016/j.molcel.2010.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley, E.D., Yeh Y.C., Hong S.-H., Fero M.J., Abeliuk E., McAdams H.H., and Shapiro L.. 2011. Assembly of the Caulobacter cell division machine. Mol. Microbiol. 80:1680–1698. 10.1111/j.1365-2958.2011.07677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, J.B., Muthusamy A.K., Liang Y., Brown T.A., Lemon W.C., Patel R., Lu R., Macklin J.J., Keller P.J., Ji N., and Lavis L.D.. 2017. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods. 14:987–994. 10.1038/nmeth.4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich, C., Templin M.F., Ursinus A., Merdanovic M., Berger J., Schwarz H., de Pedro M.A., and Höltje J.V.. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167–178. 10.1046/j.1365-2958.2001.02499.x [DOI] [PubMed] [Google Scholar]
- Käshammer, L., van den Ent F., Jeffery M., Jean N.L., Hale V.L., and Löwe J.. 2023. Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL. Nat. Microbiol. 8:1149–1159. 10.1038/s41564-023-01368-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, A., Vanhecke A., Archetti A., Holden S., Schaber F., Pincus Z., Laub M.T., Goley E., and Manley S.. 2018. Constriction rate modulation can drive cell size control and homeostasis in C. crescentus. iScience. 4:180–189. 10.1016/j.isci.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere, P.J., Mahone C.R., Santiago-Collazo G., Howell M., Daitch A.K., Zeinert R., Chien P., Brown P.J.B., and Goley E.D.. 2019. An essential regulator of bacterial division links FtsZ to cell wall synthase activation. Curr. Biol. 29:1460–1470.e4. 10.1016/j.cub.2019.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere, P.J., Szwedziak P., Mahone C.R., Löwe J., and Goley E.D.. 2018. FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division. Mol. Microbiol. 107:180–197. 10.1111/mmi.13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Boes A., Cui Y., Zhao S., Liao Q., Gong H., Breukink E., Lutkenhaus J., Terrak M., and Du S.. 2022. Identification of the potential active site of the septal peptidoglycan polymerase FtsW. PLoS Genet. 18:e1009993. 10.1371/journal.pgen.1009993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Persons L., Lee L., and de Boer P.A.J.. 2015. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 95:945–970. 10.1111/mmi.12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Sciuto, A., Fernández-Piñar R., Bertuccini L., Iosi F., Superti F., and Imperi F.. 2014. The periplasmic protein TolB as a potential drug target in Pseudomonas aeruginosa. PLoS One. 9:e103784. 10.1371/journal.pone.0103784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, S.J., Velle K.B., Mullins R.D., and Fritz-Laylin L.K.. 2020. SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219:e202001064. 10.1083/jcb.202001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu, Z., Yahashiri A., Yang X., McCausland J.W., Kaus G.M., McQuillen R., Weiss D.S., and Xiao J.. 2022. FtsN maintains active septal cell wall synthesis by forming a processive complex with the septum-specific peptidoglycan synthases in E. coli. Nat. Commun. 13:5751. 10.1038/s41467-022-33404-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahone, C.R., and Goley E.D.. 2020. Bacterial cell division at a glance. J. Cell Sci. 133:jcs237057. 10.1242/jcs.237057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmont, L.S., and Bernhardt T.G.. 2020. A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc. Natl. Acad. Sci. USA. 117:23879–23885. 10.1073/pnas.2004598117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCausland, J.W., Yang X., Squyres G.R., Lyu Z., Bruce K.E., Lamanna M.M., Söderström B., Garner E.C., Winkler M.E., Xiao J., and Liu J.. 2021. Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nat. Commun. 12:609. 10.1038/s41467-020-20873-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillen, R., and Xiao J.. 2020. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu. Rev. Biophys. 49:309–341. 10.1146/annurev-biophys-121219-081703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, E.L., Daitch A.K., Yao Q., Bhargava A., Jensen G.J., and Goley E.D.. 2017. FtsEX-mediated regulation of the final stages of cell division reveals morphogenetic plasticity in Caulobacter crescentus. PLoS Genet. 13:e1006999. 10.1371/journal.pgen.1006999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell, J.W., Hopkins A.C., and Laub M.T.. 2011. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 25:1328–1343. 10.1101/gad.2038911.a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell, J.W., Kambara T.K., Perchuk B.S., and Laub M.T.. 2014. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12:e1001977. 10.1371/journal.pbio.1001977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný, M., Křížek P., Borkovec J., Svindrych Z., and Hagen G.M.. 2014. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 30:2389–2390. 10.1093/bioinformatics/btu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, S., Jenal U., and Katayama T.. 2020. Novel divisome-associated protein spatially coupling the Z-ring to the chromosomal replication terminus in Caulobacter crescentus. MBio. 11:e00487-20. 10.1128/mBio.00487-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K.T., Du S., and Lutkenhaus J.. 2020. Essential role for ftsL in activation of septal peptidoglycan synthesis. MBio. 11:e03012–e03020. 10.1128/mBio.03012-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K.T., Pichoff S., Du S., and Lutkenhaus J.. 2021. FtsA acts through FtsW to promote cell wall synthesis during cell division in Escherichia coli. Proc. Natl. Acad. Sci. USA. 118:e2107210118. 10.1073/pnas.2107210118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, A.J., Cesbron Y., Shaw S.L., Bazan Villicana J., Tsui H.C.T., Boersma M.J., Ye Z.A., Tovpeko Y., Dekker C., Holden S., and Winkler M.E.. 2019. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA. 116:3211–3220. 10.1073/pnas.1816018116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, N.T., Dinh T., and Bernhardt T.G.. 2011. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J. Bacteriol. 193:4973–4983. 10.1128/JB.00316-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs, P.D.A., Buss J., Sim S.I., Squyres G.R., Srisuknimit V., Smith M., Cho H., Sjodt M., Kruse A.C., Garner E.C., et al. 2018. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14:e1007726. 10.1371/journal.pgen.1007726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini, I.F., and Koumoutsakos P.. 2005. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151:182–195. 10.1016/j.jsb.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Schäper, S., Brito A.D., Saraiva B.M., Squyures G.R., Holmes M.J., Garner E.C., Hensel Z., Henriques R., and Pinho M.G.. 2023. Processive movement of Staphylococcus aureus essential septal peptidoglycan synthases is independent of FtsZ treadmilling and drives cell constriction. bioRxiv. 10.1101/2023.06.29.547026 (Preprint posted June 29, 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH image to imageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler, M., Iniesta A.A., and Shapiro L.. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137. 10.1093/nar/gkm818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, M.J., and Bernhardt T.G.. 2015. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95:925–944. 10.1111/mmi.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara, T., Parzych K.R., Dinh T., and Bernhardt T.G.. 2010. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29:1412–1422. 10.1038/emboj.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling, M.C.F., and Thanbichler M.. 2020. Generating asymmetry in a changing environment: Cell cycle regulation in dimorphic alphaproteobacteria. Biol. Chem. 401:1349–1363. 10.1515/hsz-2020-0235 [DOI] [PubMed] [Google Scholar]
- Wang, L., and Lutkenhaus J.. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29:731–740. 10.1046/j.1365-2958.1998.00958.x [DOI] [PubMed] [Google Scholar]
- Wang, S.C.E., West L., and Shapiro L.. 2006. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J. Bacteriol. 188:1497–1508. 10.1128/JB.188.4.1497-1508.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, D.S. 2015. Last but not least: New insights into how FtsN triggers constriction during Escherichia coli cell division. Mol. Microbiol. 95:903–909. 10.1111/mmi.12925 [DOI] [PubMed] [Google Scholar]
- Whitley, K.D., Grimshaw J., Karinou E., Stansfeld P.J., and Holden S.. 2023. A one-track model for spatiotemporal coordination of Bacillus subtilis septal cell wall synthesis. bioRxiv. 10.1101/2023.06.29.547024 (Preprint posted June 29, 2023). [DOI] [Google Scholar]
- Woldemeskel, S.A., McQuillen R., Hessel A.M., Xiao J., and Goley E.D.. 2017. A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus. Mol. Microbiol. 105:721–740. 10.1111/mmi.13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahashiri, A., Jorgenson M.A., and Weiss D.S.. 2015. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl. Acad. Sci. USA. 112:11347–11352. 10.1073/pnas.1508536112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., McCausland J.W., Lyu Z., and Xiao J.. 2023. Scripts for analyzing single-molecule tracking trajectories in bacteria. Zenodo. 10.5281/zenodo.4306645 [DOI] [Google Scholar]
- Yang, X., and Liu R.. 2022. How does FtsZ’s treadmilling help bacterial cells divide? Biocell. 46:2343–2351. 10.32604/biocell.2022.022100 [DOI] [Google Scholar]
- Yang, X., Lyu Z., Miguel A., McQuillen R., Huang K.C., and Xiao J.. 2017. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 355:744–747. 10.1126/science.aak9995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., McQuillen R., Lyu Z., Phillips-Mason P., De La Cruz A., McCausland J.W., Liang H., DeMeester K.E., Santiago C.C., Grimes C.L., et al. 2021. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. 6:584–593. 10.1038/s41564-020-00853-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X.C., Weihe E.K., and Margolin W.. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 180:6424–6428. 10.1128/jb.180.23.6424-6428.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the summary of SMT of Halo-FtsW and Halo-FzlA variants with varying states of activation.
shows proteins identified by MS that are fivefold enriched in relative abundance in 3xFLAG-FzlA eluate compared to WT.
shows the summary of DNA damage reporter experiments.
shows strains and plasmids used in this study.
is the source file for Fig. 7.
is the source file for Fig. S1.
Data Availability Statement
Data are available in the article itself and its supplementary materials. Strains and plasmids generated in this study are available from the corresponding author upon request.