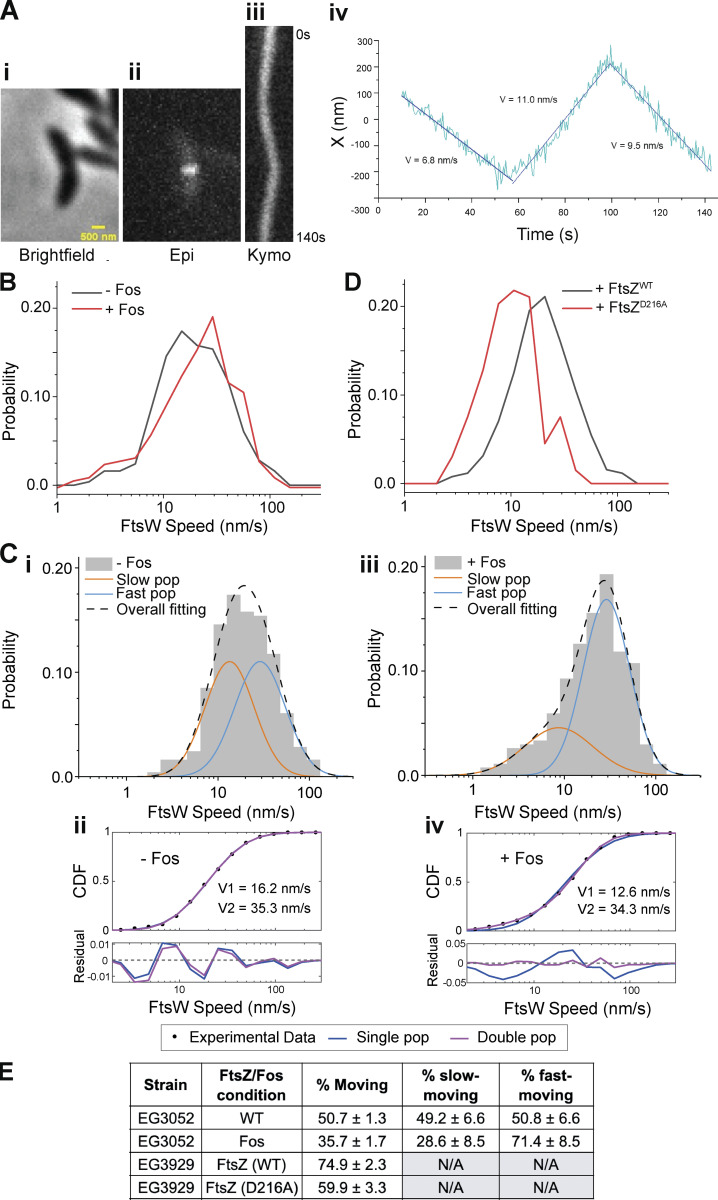
Figure 2.
FtsW in Caulobacter moves in two populations. (A i) Brightfield image of a Halo-ftsW**I* cell. (ii) Representative maximum fluorescence intensity projection (Epi) for Halo-FtsW**. (iii) Kymograph (Kymo) of the fluorescence signal of a line scan across the division plane for a Janelia Fluor 646–labeled single Halo-FtsW** (EG3053). (iv) Plot of molecule position at midcell over the course of imaging and speeds of movement for each segment. X indicates the short axis of the cell. (B) Speed distribution of directionally moving Halo-FtsW (EG3052) molecules with (n = 318 from four biological replicates) and without (n = 256 total from three biological replicates) Fos treatment. (C i and iii) Two-population fitting of the distributions in B without (i) or with (iii) Fos treatment. Orange and blue curves represent the slow- and fast-moving populations, respectively. The black dashed curve is the overall fit of the distribution. (ii and iv) Goodness of fit (top) of two- (magenta) versus one- (blue) population fitting of Halo-FtsW speed distribution. The residuals of each fit (bottom) illustrate how well each model captures the data at every point along the CDF. Data in C is duplicated in Fig. 3 B. (D) Speed distribution of directionally moving Halo-FtsW molecules in cells (EG3929) producing FtsZWT (n = 254 total from three biological replicates) or FtsZD216A (n = 131 total from three biological replicates). (E) Fraction of the FtsW population that is moving overall (% moving), slow moving, or fast moving for the indicated strains and conditions.