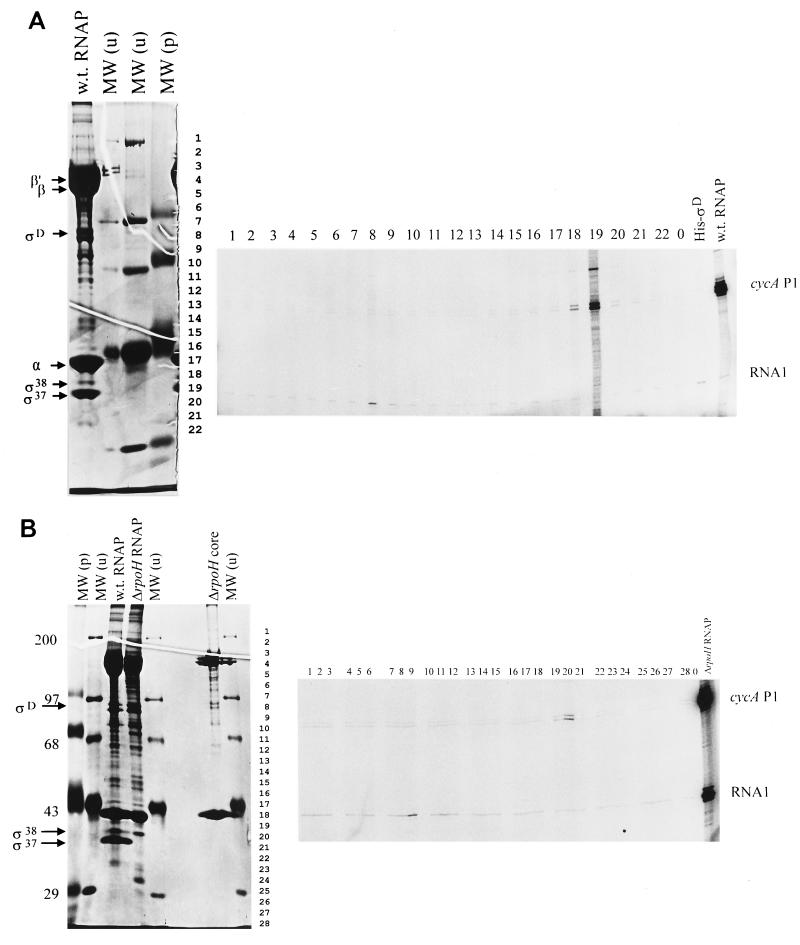
FIG. 4.
Transcription of the G34T mutant promoter by reconstituted RNA polymerase holoenzymes. (A) Transcripts produced when the G34T promoter is incubated with different R. sphaeroides RNA polymerase samples. The positions of transcripts are indicated to the right of the rightmost gels as follows: RNA1, the EςD-dependent oriV transcript from the transcription plasmid template (10); cycA P1, the product of the G34T promoter. Reactions in panel A used renatured proteins from a wild-type RNA polymerase sample (shown on the left) as a source of potential sigma factors for addition to core RNA polymerase (shown in panel B) from the ΔRpoH mutant (11). Reactions in panel B used renatured proteins from a RpoH null mutant as a source of potential sigma factors (11) for reconstitution with the same ΔRpoH core RNA polymerase. In panels A and B, numbers above the rightmost gels denote the SDS-polyacrylamide gel fraction that was added to core RNA polymerase as a source of potential sigma factor. Lane 0 contains transcripts produced by core RNA polymerase alone. Lane His-ςD in panel A shows transcripts produced when recombinant R. sphaeroides His-ςD is added to core RNA polymerase. To the left of the leftmost gels in panels A and B is shown the polypeptide composition of RNA polymerase samples used for in vitro transcription reactions. These samples include RNA polymerase samples from wild-type cells (w.t. RNAP in panels A and B) or a ΔRpoH mutant (11) (ΔrpoH RNAP in panel B) and core RNA polymerase from an RpoH null strain (ΔrpoH core in panel B) separated alongside prestained [MW (p)] or unstained [MW (u)] protein molecular weight standards (molecular weights [in thousands] indicated on the left margin). Numbers to the right of the leftmost gel indicate how this gel was sliced to obtain proteins to be tested for sigma factor activity. Individual RNA polymerase subunits are indicated to the left.