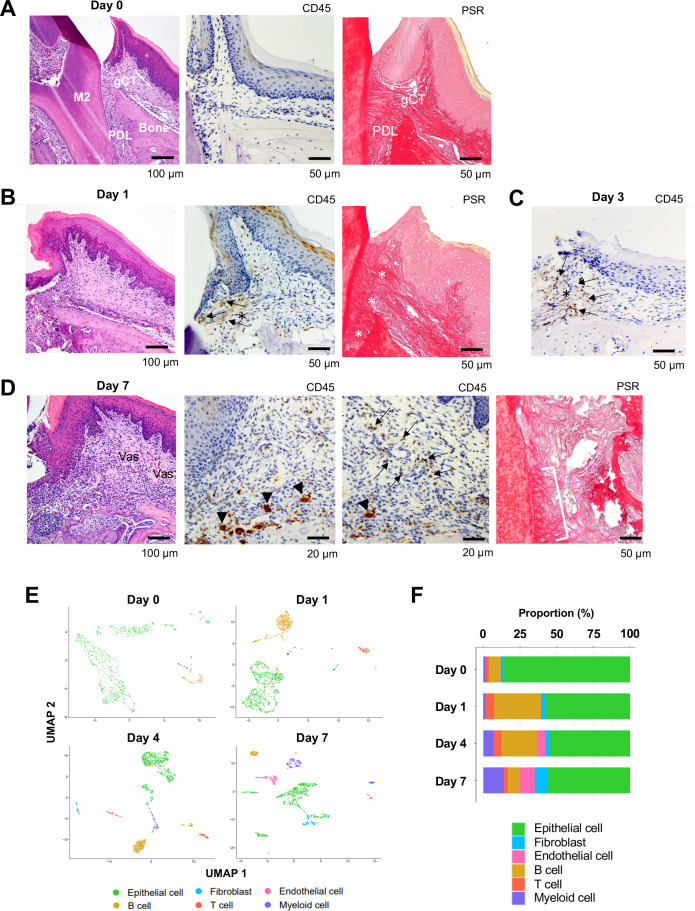
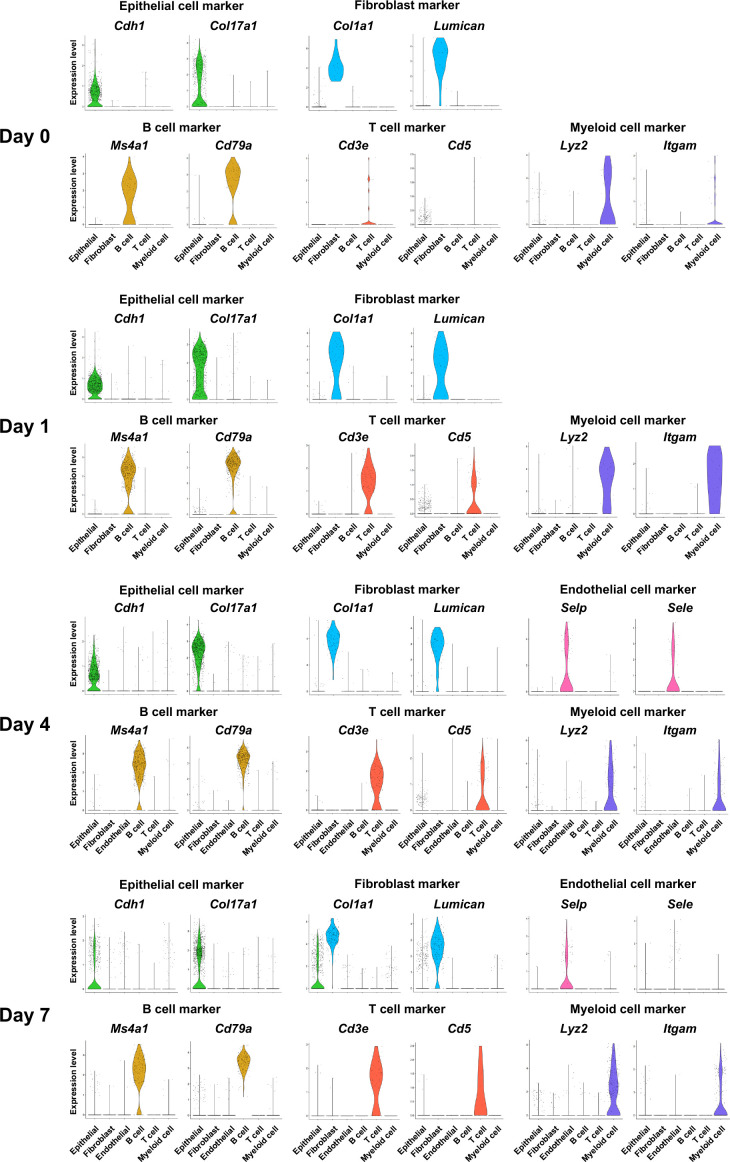
Figure 1. Changes in proportions of major cell types during ligature-induced periodontitis development in mice.
(A) Mouse maxillary second molar (M2) has multi-roots and supported by alveolar bone (Bone), periodontal ligament (PDL), and gingiva connective tissue (gCT). The oral barrier immunity is not constitutively activated as evidenced by the lack of CD45+ immune cells. Picrosirius red (PSR)-stained collagen fibers connected the root surface and alveolar bone in the PDL and organized as dense parallel bundles in gCT. (B) One day (day 1) after a ligature (5.0 silk suture) was placed around M2, the cervical PDL and gCT demonstrated a localized connective tissue degradation (*), where CD45+ immune cells clustered (arrows). PSR-stained collagen architecture immediately under the ligature lost the thick collagen bundle structure (*). (C) Day 3 of ligature placement exhibited localized but increased CD45+ immune cell clustering adjacent to the collagen degradation area (*). (D) Day 7 of ligature placement, PDL, and gCT tissue degradation increased with inflammatory vascularization (Vas). CD45+ myeloid cells (arrowheads) were observed near the alveolar bone surface and CD45+ lymphocytes (arrows) infiltrated the gCT area. PSR staining lost the typical collagen pattern in gCT and PDL, and a remnant of degraded PDL collagen fiber (white bracket) was attached to the tooth surface. (E) Single-cell RNA sequencing (scRNA-seq) t-distributed stochastic neighbor embedding (t-SNE) projection plots showing the major cell types present within gingival tissue during periodontitis development on days 0, 1, 4, and 7. Colors indicate cell type as follows: green, epithelial cells; blue, fibroblasts; pink, endothelial cells; yellow, B cells; red, T cells; and purple, myeloid cells. (F) Proportion plots showing the relative amounts of each major cell type on days 0, 1, 4, and 7.