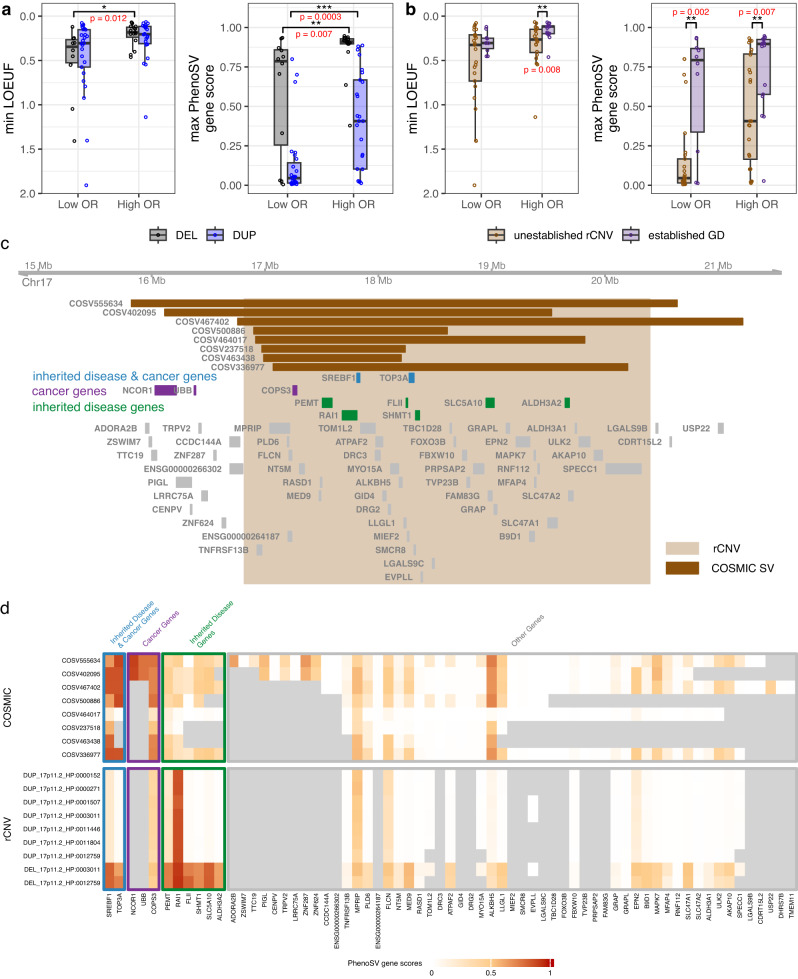
Fig. 5. PhenoSV predictions aid in identifying disease-related genes from SVs that are implicated in distinct phenotypes.
a Comparisons of genes covered by incompletely penetrant rCNV segments in the bottom third odds ratios (Low OR, n = 12 deletions and n = 27 duplications) and highly penetrant rCNV segments of the top third odds ratios (High OR, n = 16 deletions and n = 25 duplications). b Comparisons of genes covered by incompletely penetrant rCNV segments in the bottom third odds ratios (Low OR, 29 unestablished rCNV and 10 established GD) and highly penetrant rCNV segments of the top third odds ratios (High OR, n = 24 unestablished rCNV and n = 17 established GD). a, b Gene constraints were measured by minimum LOEUF (left panel) scores and maximum scores (right panel) of genes affected by each rCNV segment. Median (center line), IQR (box limits), and outliers (points) that exceeding 1.5x IQR were shown in the boxplot. Two-sided Wilcoxon rank sum test p value < 0.05*, <0.01**, <0.001***. c Displayed is an example of overlapping rCNV (chr17:16816686-20396687) and 8 COSMIC SVs affecting similar sets of genes, including genes associated with both inherited diseases and cancers, genes primarily associated with inherited diseases, genes mainly associated with cancers, and other genes. d Heatmap of gene-level PhenoSV predictions () for 8 COSMIC SVs (upper panel) and 9 rCNV-phenotype pairs from (c). Genes associated with both inherited diseases and cancers, and genes mainly associated with cancers were given high scores for COSMIC SVs. Genes associated with both inherited diseases and cancers, and genes mainly associated with inherited diseases were given high scores for rCNVs. Source data are provided as a Source Data file. LOEUF loss-of-function observed/expected upper bound fraction, GD genomic disorder.