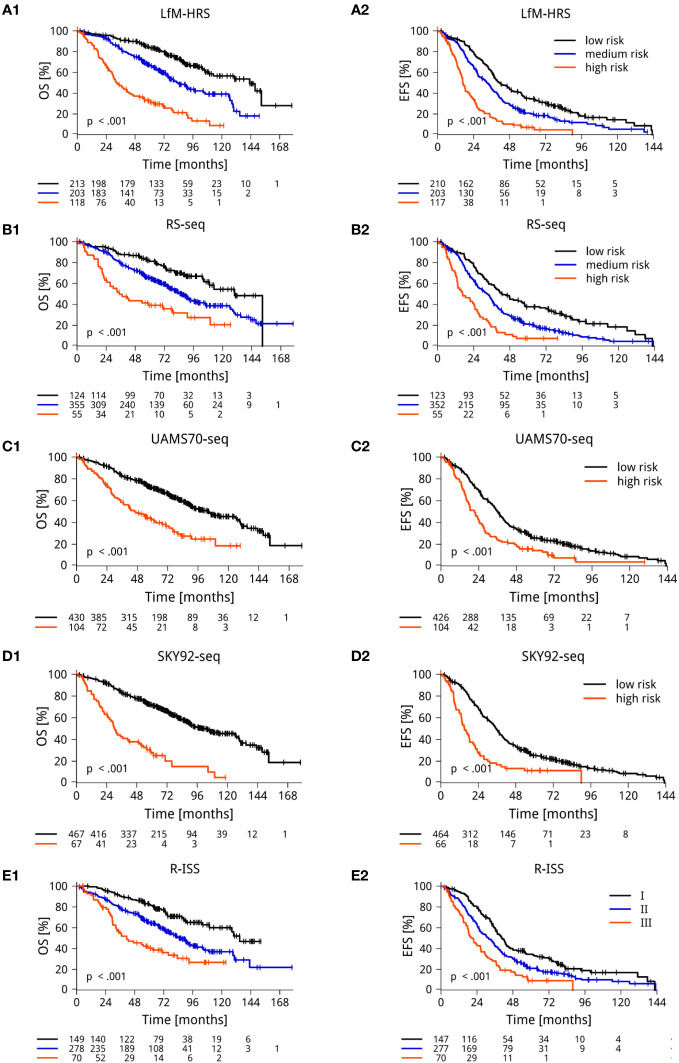
Figure 2.
RNA-sequencing based determination of risk. (A) De novo generated RNA-sequencing-based scores for risk (LfM-HRS) delineates 3 groups with significantly different overall (OS) (A1) and event-free (EFS) (A2) survival. (B-D) “GEP”-scores translated into RNA-sequencing. The scores of the Universities of Heidelberg and Montpellier (RS-score) (B), the University of Arkansas Medical School (UAMS70) (C), and the Erasmus Medical Center (SKY92), (D) in each case delineate symptomatic myeloma patients with significantly different EFS and OS. (E). The current clinical gold standard (revised ISS-score) delineates three groups of 30%, 56% and 14% of 535 patients with significantly different OS (E1) and EFS (E2). Depicted are Kaplan Maier curves with log-rank based P-value and patients at risk. P-values were adjusted for multiple testing using Benjamini-Hochberg correction. For validation of RNA-sequencing based scores on the independent CoMMpass-cohort, see Supplementary Figure S5.