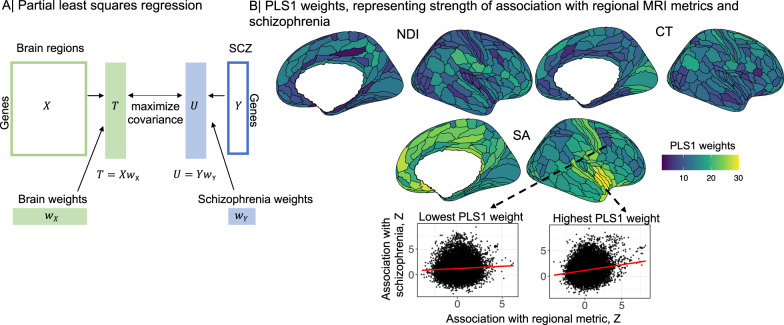
Fig. 2. Partial least squares (PLS) analysis of genetic covariation between regional brain phenotypes and schizophrenia.
A The {1 × 18,640 } vector of unthresholded gene association statistics (Z-scores) derived by H-MAGMA analysis of the schizophrenia GWAS dataset was designated as the response variable, i.e., the dependent Y vector; and the {180 × 18,640} matrix of unthresholded gene association Z-scores for each of the MRI GWAS datasets was designated the predictor variable, i.e., the independent X matrix. The first PLS component (PLS1) defined the weighted functions of X and Y that were most strongly correlated overall weighted functions of the whole genome. The PLS1 weights for X (brain weights, w(X)i, i = 1, 2, 3, …180) multiplied by X constituted a {1 × 18,640} vector of T scores (genes weighted by association with brain phenotypes); whereas, the PLS1 weights for Y (schizophrenia weights w(Y)i) multiplied by Y constituted a {1 × 18,640} vector of U scores (genes weighted by association with schizophrenia). Thus genes with the highest absolute T and U scores can be regarded as the genes which contribute most strongly to the genetic covariation between schizophrenia and each regional brain phenotype32,108. B Cortical surface maps of PLS1 weights for neurite density index (NDI), cortical thickness (CT) and surface area (SA). Cortical regions with higher PLS1 weights (shades of yellow) have stronger genetic covariation with schizophrenia: for SA, regions of insular and medial prefrontal cortex; for CT, visual, premotor and inferior parietal cortex; and for NDI, inferior frontal, inferior parietal, posterior cingulate and posterior opercular cortex. Scatterplots (Spearman’s correlations, ρ) illustrate the genetic relationships between schizophrenia (y-axis, Z-scores from H-MAGMA analysis of schizophrenia GWAS dataset) and brain surface area (x-axis, Z-scores from H-MAGMA analysis of MRI GWAS datasets) in two cortical regions, one with a low PLS1 weight (left, dark blue, ρ = 0.04), and one with a high PLS1 weight (right, yellow, ρ = 0.17). In both plots each point represents one of 18,640 genes. Spearman’s correlations were two-tailed. Source data are provided as a Source Data file.