Abstract
We have isolated a UV-induced temperature-sensitive mutant of Mycobacterium smegmatis that fails to grow at 42°C and exhibits a filamentous phenotype following incubation at the nonpermissive temperature, reminiscent of a defect in cell division. Complementation of this mutant with an M. smegmatis genomic library and subsequent subcloning reveal that the defect lies within the M. smegmatis dnaG gene encoding DNA primase. Sequence analysis of the mutant dnaG allele reveals a substitution of proline for alanine at position 496. Thus, dnaG is an essential gene in M. smegmatis, and DNA replication and cell division are coupled processes in this species. Characterization of the sequences flanking the M. smegmatis dnaG gene shows that it is not part of the highly conserved macromolecular synthesis operon present in other eubacterial species but is part of an operon with a dgt gene encoding dGTPase. The organization of this operon is conserved in Mycobacterium tuberculosis and Mycobacterium leprae, suggesting that regulation of DNA replication, transcription, and translation may be coordinated differently in the mycobacteria than in other bacteria.
Mycobacteria are distinct from other organisms due to several unique physical characteristics. One of these characteristics is a thick, mycolic acid-rich cell wall which differs substantially from those of gram-negative and gram-positive organisms (6). The second striking feature is an extremely low growth rate. Mycobacteria can be classified broadly into two groups based on growth rate; fast-growing species such as Mycobacterium smegmatis have a doubling time of 3 to 4 h, while slow-growing species such as Mycobacterium tuberculosis have a doubling time of 24 h (53). Mycobacterium leprae has an even slower doubling time, determined to be 13 days in mice (23, 24). The vast majority of pathogenic strains are slow growing, while the fast-growing strains are mostly nonpathogenic, saprophytic organisms. Even the fast-growing mycobacterial species have doubling times which are six times slower than that of the well-characterized Escherichia coli under similar conditions. Thus, it is likely that physiological processes such as DNA replication, transcription, and cell division are regulated differently in these unusually slow-growing microorganisms. Specifically, DNA replication and cell division have been shown to be linked in many other prokaryotes through the characterization of DNA replication mutants. The dnaG-encoded primase has been shown to be an essential DNA replication protein in E. coli, and mutants in primase often exhibit a filamentous phenotype indicative of the linkage between DNA replication and cell division (52).
The characterization of temperature-sensitive E. coli mutants has provided a wealth of knowledge about the essential role of the dnaG-encoded primase in the process of DNA replication in E. coli. In E. coli, the dnaG gene has been shown to lie in an operon at 67 min, consisting of the genes rpsU, dnaG, and rpoD (9, 29, 30). The dnaG gene encodes the primase protein, an essential enzyme that recognizes specific nucleotide sequences and then synthesizes small primer RNAs which act as a substrate for the initiation of DNA replication (5, 21, 48). The rpoD gene codes for the ς subunit of the RNA polymerase and is responsible for recognition of the promoter sequences and the subsequent initiation of transcription (9, 54). The rpsU gene encodes the S21 protein which enables the 3′ end of the 16S rRNA to recognize the ribosome binding site (41). A number of regulatory features have been identified in this operon, including seven promoters, two transcriptional terminators, the nut equivalent pN-like binding site, and a potential LexA binding site (9, 25, 26, 28, 30, 42). The inclusion of genes involved in regulating the initiation of synthesis of the major informational macromolecules of the cell prompted researchers to name this operon the macromolecular synthesis operon (26).
This gene organization suggests that under certain physiological conditions there may be a need for coregulation of the expression of major macromolecules in the cell (26, 30). It appears that there has been strong pressure to conserve the organization of this operon, since it has been identified in at least six diverse bacterial species, including Listeria monocytogenes, Lactococcus lactis, Bacillus subtilis, Haemophilus influenza, Rickettsia prowazekii, and Myxococcus xanthus (49, 50), and was thought to possibly be universally conserved (26).
Very little is known about the proteins involved in DNA replication in mycobacteria, and there have been no well-characterized conditional-lethal mycobacterial DNA replication mutants. It is extremely difficult to obtain temperature-sensitive mutants in mycobacteria, which are essential to study processes such as DNA replication and cell division. In this study, we describe conditional-lethal M. smegmatis mutants in dnaG and the novel organization of the dnaG locus of M. smegmatis, which is apparently conserved in slow- and fast-growing mycobacteria. Interestingly, these data show the physiological consequences of a defect in DNA replication in M. smegmatis as well as the novel organization of the macromolecular synthesis operon in mycobacteria. The fact that the mycobacteria have a different organization of the genes comprising the dnaG locus suggests that they exhibit a significant departure from the way other bacteria coordinate the synthesis of their macromolecules.
MATERIALS AND METHODS
Bacterial strains and plasmids.
M. smegmatis mc2155 (43, 44) was used in all of the mutagenesis experiments and has been described previously. The pYUB415-based cosmid libraries of M. smegmatis and Mycobacterium bovis BCG used for complementation experiments were a kind gift from W. R. Jacobs, Jr. The plasmid pUC119 (38) and the E. coli-mycobacterial shuttle plasmid vector pMD31 (13) have been described previously.
Growth of bacterial cultures.
M. smegmatis cultures were grown in Middlebrook 7H9 broth (Difco, Detroit, Mich.) supplemented with 4% glucose and 0.2% Tween 80 (Difco). Agar plates were prepared with the same supplements plus 50 μg of carbenicillin per ml and 10 μg of cycloheximide per ml. E. coli cultures were grown in Luria broth.
Isolation and characterization of mutants.
A germicidal UV lamp was used at a strength of 600 μJ/cm2 16.5 cm from the cultures. Optical densities (OD) of the cultures were taken with a lambda (Perkin-Elmer Corp., Norwalk, Conn.) 3.3 UV/vis spectrophotometer at a wavelength of 600 nm. A modification of the method of Miller (32) was used for the mutagenesis procedure. Cells were diluted to an optical density at 600 nm (OD600) of 0.1 to 0.2 (which corresponds to approximately 107 CFU/ml). A killing curve was established for strain mc2155 by placing a 500-μl sample of the mc2155 culture on an uncovered petri dish and exposing it to the UV light for various intervals, after which a 100-μl aliquot was removed to a tube containing 400 μl of fresh broth. Only two aliquots were removed from each 500-μl sample to ensure uniform exposure of the culture to the mutagen. The time intervals tested were 0 and 15 s, 30 and 45 s, and 60 and 90 s. The mutagenized culture was allowed to recover in the dark at 30°C for 4 h, at which time dilutions were plated and incubated for 2 days. These colonies were then patched in duplicate onto gridded plates and incubated at the permissive temperature, 30°C, and the nonpermissive temperature of 42°C. Mutant colonies failed to grow at the nonpermissive temperature and were grown from the permissive cultures and further characterized. The mutants described here were isolated from a 30-s exposure in which there was approximately 60% killing of the cells. The mutants were named SMEG 1, SMEG 2, etc. (for M. smegmatis mutants in essential genes).
Staining of cells.
Acid-fast staining of the cells was performed with the BBL TB Kinyoun stain kit (Becton Dickinson, Raleigh-Durham, N.C.) as previously described, with heating (19). DAPI (4′, 6-diamidino-2-phenylindole) staining was performed according to the method of Grompe et al. (14) with the following changes. The M. smegmatis strains were grown to an OD600 of 1.0 and were then washed and resuspended in a 0.84% NaCl solution. Twenty microliters of cells were applied to a glass slide, air dried, and fixed with methanol. Ten microliters of a poly-l-lysine solution (5 μg/ml) (Sigma) was applied to the slide and allowed to dry. Ten microliters of DAPI stain (5 μg/ml; Boehringer Mannheim, Indianapolis, Ind.) was applied to the slide and allowed to air dry. A coverslip was placed on the sample area, and the edges were sealed with clear nail polish. The slides were examined by fluorescence and phase-contrast microscopy.
Microscopy.
Wild-type and mutant cells were visualized with a Zeiss phase-contrast microscope with 20 μl of sample applied to a clean slide topped by a Corning no. 1 glass coverslip. Photographs were taken at a magnification of 1,000× with a Nikon camera and Kodak Ektachrome 160T film (Kodak, New Haven, Conn.).
Recombinant plasmids.
All enzymatic DNA manipulations were performed according to the manufacturers’ instructions (Boehringer Mannheim and New England BioLabs). The pYUB415::mc2155 cosmid library (a kind gift from W. R. Jacobs, Jr.) is similar to others previously described (17, 18, 35, 39). This library contains genomic DNA fragments approximately 30 kb in size cloned into the E. coli-mycobacterial shuttle cosmid vector pYUB415 (encoding carbenicillin resistance and hygromycin resistance).
Complementation and subcloning of the mutants.
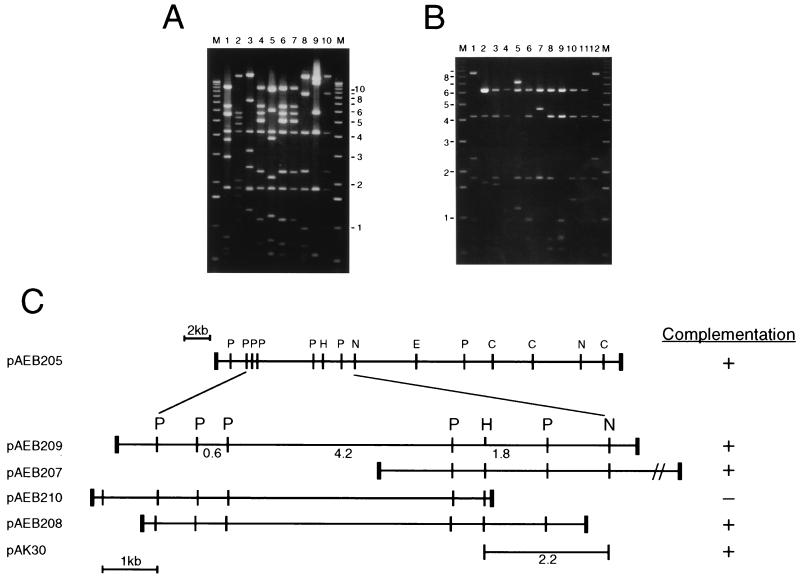
Mapping of all 10 complementing cosmids showed that they contain three PvuII fragments (4.2, 1.8, and 0.6 kb) in common (Fig. 2), although other restriction fragments are not shared among subsets of the cosmids, so it was not possible to position the cosmids with respect to each other. The insert DNA in pAEB205 was isolated and used as a probe against a PvuII digest of all 10 cosmids and M. smegmatis chromosomal DNA, which indicated that some of these cosmids appeared to have undergone rearrangements; however, the hybridization patterns of pAEB205 and the M. smegmatis chromosome are very similar, suggesting that pAEB205 is not rearranged (data not shown).
FIG. 2.
Complementing cosmids and plasmids of SMEG 23. (A) Agarose gel electrophoresis of PvuII digestions of 10 cosmids recovered from temperature-resistant transformants of SMEG 23. Cosmid pAEB205 is shown in lane 8. Sizes of DNA markers (M) are shown in kilobase pairs. (B) Agarose gel electrophoresis of PvuII digestions of plasmid subclones derived from cosmid pAEB205 DNA. Plasmids pAEB209, pAEB207, pAEB210, and pAEB208 are shown in lanes 2, 5, 8, and 9, respectively. (C) Restriction map of cosmid pAEB205 and plasmids derived from it with their corresponding complementation of SMEG 23. Relevant restriction sites for PvuII (P), EcoRV (E), ClaI (C), HindIII (H), and NheI (N) are shown. The positions of the 0.6-, 4.2-, and 1.8-kb PvuII fragments are shown. Thick vertical bars indicate the junctions between the insert DNA and vector DNA (not shown).
To facilitate further mapping of the complementing region, cosmid pAEB205 was partially digested with Sau3AI, and 7- to 12-kb fragments were inserted into the shuttle vector pMD30 (13). Approximately 200 recombinant clones were screened by hybridization with the 4.2-kb PvuII fragment common to all the complementing cosmids, and 10 positively hybridizing plasmids, each of which was found to contain at least two of the three common PvuII fragments, were identified (Fig. 2B). Plasmid pAK30 was constructed by ligating the HindIII-NheI fragment containing the wild-type dnaG gene to the HindIII and XhoI sites of pMD31. DNA fragment purification was done with the Elutrap system (Schleicher & Schuell, Keene, N.H.).
Transformations and transfections.
The introduction of plasmid DNA into E. coli was performed according to standard procedures (38) or by electroduction from M. smegmatis cells (2). Plasmid DNAs were introduced into M. smegmatis mc2155 and mutant strains by electroporation (43), recovered for 2 h, and plated directly onto selective media. A standard procedure for lambda infection (31) was used to transfect DH5α with the pYUB415::mc2155 cosmid-library lysate. Plates containing approximately 106 transfectants were harvested; cosmid DNAs were isolated by alkaline lysis (38).
DNA hybridization.
M. smegmatis genomic DNA was prepared by a method that was similar to those previously described (22). Southern hybridizations were performed following the transfer of DNA to a Nytran Plus membrane (Schleicher & Schuell). DNA labeling and detection were performed by the Genius system (Boehringer Mannheim).
DNA sequencing and analysis.
A 5,168-bp fragment from subclone 15 was sequenced with a redundancy of 8 with a Perkin-Elmer ABI Prism 377 automated sequencer. Samples were prepared for cycle sequencing according to the manufacturer’s instructions. Plasmids for sequencing were prepared by standard procedures with DNase I digestions generating several hundred 1- to 3-kb constructs which were ligated to the pBluescript KS vector and sequenced with forward and reverse primers as well as with sequence-specific primers. G+C content was determined to be 67% for this region. The sequence was compiled by the Sequencher program (Gene Codes Corp., Ann Arbor, Mich.) with a Power MacIntosh 7100/66. Analyseq, Genemark, Beauty, and the Genetics Computer Group, University of Wisconsin package were all used in sequence analysis (14, 11, 45, 55). The mutation in SMEG 1, SMEG 15, SMEG 21, and SMEG 23 was determined by sequencing 1- to 2-kb regions of PCR-amplified genomic DNA. Four sets of primers were used, and multiple PCRs were sequenced to verify the fidelity of the thermostable polymerase. The primers used for amplification and sequencing were p1, 5′ GTCCACCACCAACGTGC 3′; pA, 5′ CGCGTGGTCATCGAC 3′; pC, 5′ CGTTGACCAGGTTGG 3′; pall3, 5′ GCGCGCGCCATATGGTATCCACGAGGTCG 3′; pall4, 5′ GCGCGGATCCCAACGGCACAACAGTGTCAC 3′; p22, 5′ TGA CCGTCGAGAGCTTCAC 3′; and p23, 5′ GTCAACGGCACAACAGTG 3′.
Nucleotide sequence accession number.
The sequence of the dnaG gene has been deposited in GenBank under accession no. AF 027507.
RESULTS
Isolation and identification of temperature-sensitive M. smegmatis mutants.
Temperature-sensitive mutants of M. smegmatis were isolated by exposing cultures to UV radiation as described in Materials and Methods. The cultures were allowed to recover and were then incubated on plates at the permissive temperature of 30°C for 3 days. The colonies were patched in duplicate and incubated at 30°C as well as at the nonpermissive temperature of 42°C. Cells which failed to grow at 42°C were cultured at 30°C and further analyzed. Twenty-four mutants were isolated in this manner. The mutants were determined to be sensitive to infection by the mycobacteriophages TM4, D29, and L5 (data not shown). Four mutants, SMEG 1, SMEG 15, SMEG 21, and SMEG 23, were selected for further characterization based on their phenotypic properties at the nonpermissive temperature, although they are extremely similar and are probably siblings (see below).
Physical characterization of SMEG 1.
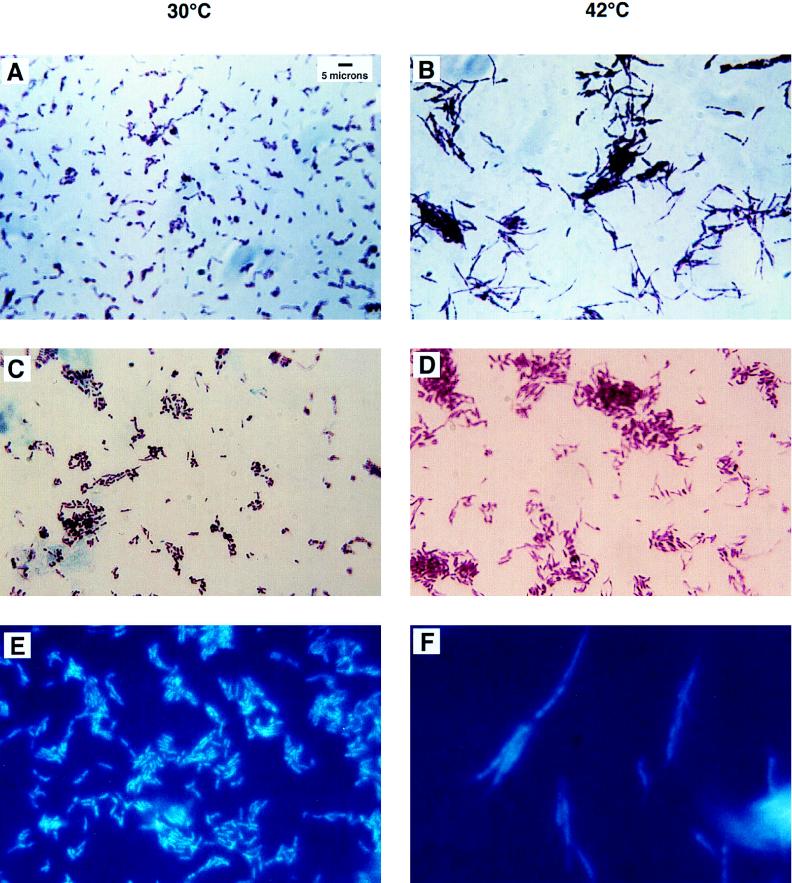
Samples of mc2155 and SMEG 1 cells were subjected to acid-fast staining after growth at the permissive and nonpermissive temperatures and were analyzed by light microscopy. Cultures were grown overnight at 30°C to an OD600 of 1.0 and were then divided; half the culture was incubated at 30°C, and the remainder was incubated at 42°C. Aliquots of 0.1 ml were taken at 2-h intervals from each culture, and bacteria were acid-fast stained and analyzed for any changes in size or shape of the individual cells by light microscopy. Mutant SMEG 1 and mc2155 cells taken from cultures incubated at the permissive temperature were indistinguishable at all time points. Additionally, there were no obvious differences detected in cultures incubated at the nonpermissive temperature until after 6 h of incubation, when the SMEG 1 mutant cells appeared elongated and filamentous (Fig. 1). These mutant cells appeared to average 2 to 3 times the length of the wild-type cells, which is generally 2 to 3 μm (20).
FIG. 1.
Acid-fast stains of SMEG 1 after growth at the permissive and nonpermissive temperatures for 6 h. (A) SMEG 1 cells incubated at 30°C appear as normal bacilli which average 2 to 3 μm in length. (B) SMEG 1 cells appear long and filamentous after 6 h of incubation at 42°C. (C) SMEG 1(pAK30) at 30°C shows the wild-type phenotype with additional copies of dnaG provided on an extrachromosomal plasmid. (D) SMEG 1(pAK30) at 42°C exhibits a wild-type phenotype showing that it has been rescued by the wild-type copy of dnaG provided by the plasmid pAK30. (E) SMEG 1 cells were incubated at 30°C and then stained with DAPI. These stained cells show nucleoids that are stained uniformly and are condensed and centrally located. (F) SMEG 1 cells after 6 h of incubation at 42°C have faintly stained, poorly defined, diffuse nucleiods.
SMEG 1 and mc2155 cells were stained with a DNA-specific dye, DAPI. This stain revealed the diffuse, irregular nature of the nucleoid structures in the SMEG 1 cells after incubation at 42°C for 6 h, while the SMEG 1 cells (as well as the mc2155 cells at both temperatures) at the permissive temperature had more distinct, condensed nucleiods (Fig. 1). The mutant phenotype of SMEG 1 is reminiscent of the parB and dnaG2903 mutants of E. coli described by Wada and Yura (52) and Grompe et al. (14) which arise from amino acid substitutions in the carboxy terminus of DNA primase and exhibit abnormal nucleiod formation. The appearance of the filamentous phenotype in SMEG 1 was concomitant with the strain’s inability to recover when it was returned to the permissive temperature (data not shown). SMEG 1 is able to recover when it is incubated at 42°C for up to 6 h and is then allowed to recover at 30°C on plates for 24 h. After an incubation of 6.5 h at 42°C, the cells are no longer viable and cannot grow when plated at 30°C. SMEG 15, SMEG 21, and SMEG 23 exhibit identical phenotypes.
Complementation of SMEG 1, SMEG 15, SMEG 21, and SMEG 23.
The temperature-sensitive mutant SMEG 23 was electroporated with a genomic library of M. smegmatis mc2155 DNA in the shuttle phasmid vector pYUB415, and portions of the transformation mixture were plated out at 30 and 42°C. A complementation frequency of 1.5% was observed for SMEG 23, and similar frequencies were observed for SMEG 1, SMEG 15, and SMEG 21. Cosmid DNAs from 10 of the SMEG 23 clones growing at 42°C were recovered in E. coli by electroduction (2) and digested with PvuII, showing that at least seven different cosmids were represented (Fig. 2A). One of these cosmids, pAEB205, was reintroduced into SMEG 23 and shown to complement with a 100% frequency. Cosmid DNAs were similarly recovered from SMEG 1, SMEG 15, and SMEG 21 and were shown to cross-complement all of these mutants, indicating that these are sibling derivatives or that they contain closely linked mutations. We were also able to isolate complementing cosmids from a library containing DNA from M. bovis BCG, and five such cosmids complement all of these mutants.
The dnaG gene of M. smegmatis complements SMEG 1.
The complementing gene present on the cosmid DNAs was identified by constructing a series of subclones containing smaller segments of DNA (Fig. 2). Two subclones, pAEB208 and pAK30, containing 5.1- and 2.2-kb fragments, respectively, define the minimal DNA segment able to complement SMEG 1. A 5,168-bp segment of M. smegmatis DNA derived from pAEB205 was sequenced, and five open reading frames, only one of which is present in both the pAEB208 and pAK30 subclones, were identified (Fig. 2C). This 1,908-bp open reading frame is predicted to encode a 636-amino-acid protein (69.8 kDa). Comparison of this protein sequence with the protein databases revealed that it is closely related to bacterial DNA primases encoded by dnaG genes; these searches also identified the dnaG homologs of M. tuberculosis and M. leprae (3). The DnaG proteins of M. smegmatis, M. tuberculosis, and M. leprae are very similar, and the M. smegmatis and M. tuberculosis primases have 74% identity, M. smegmatis and M. leprae have 81% and M. leprae and M. tuberculosis have 79%.
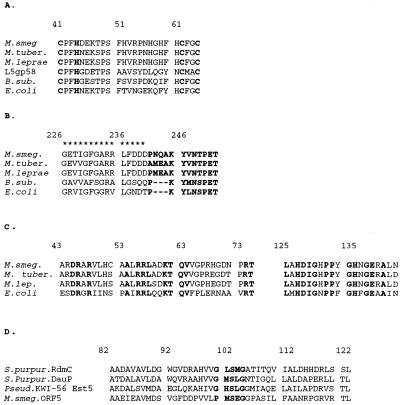
There are several highly conserved motifs and functional domains in bacterial primases, one of which is the distinctive zinc finger catalytic domain, which lies in the N terminus of the protein (16, 46). This region, as well as the bacterial dnaG signature sequence and the RNA polymerase large-subunit motif, is conserved in the M. smegmatis primase (33, 49). A partial alignment of the conserved primase domains, shown in Fig. 4, includes the primases from E. coli (GenBank no. J01687) and B. subtilis (GenBank no. M10040) for comparison.
FIG. 4.
Alignments of homologous regions of the M. smegmatis dnaG locus. (A) The putative Zn finger catalytic region of dnaG-encoded primase from E. coli, B. subtilis, M. smegmatis, M. tuberculosis, and M. leprae. Included in this alignment is mycobacteriophage L5 gp 58, which encodes the putative phage-encoded primase. The residues which may be involved in zinc binding are in bold. (B) Alignment of the region containing the RNAP-basic sequence, delineated with asterisks, as well as the bacterial dnaG signature sequence (50), in bold. (C) Alignment of an amino-terminal region of the dGTPase from E. coli with the amino-terminal region of the putative dGTPases from M. smegmatis, M. tuberculosis, and M. leprae. These regions exhibit the highest degree of similarity between the proteins in these four strains, with the region encompassing amino acids 43 to 77 exhibiting 38% identity and that from 125 to 143 showing 67% identity. The completely conserved amino acids are in bold. (D) Alignment of the two highly conserved regions of the methyltransferases RdmC and DauP from S. purpurascens as well as the esterase Est5 from Pseudomonas sp. strain KWI-56 with the partial ORF5 from M. smegmatis. The serine active site box (40) is in bold. These conserved residues are thought to be involved in forming the catalytic triad necessary for the function of these enzymes.
Identification of the mutant allele in SMEG 1.
Since the M. smegmatis dnaG gene complements SMEG 1, SMEG 15, SMEG 21, and SMEG 23, it is likely that the temperature-sensitive phenotype of these mutants is a manifestation of a mutation in their dnaG genes. This possibility was confirmed by PCR amplification of a DNA segment containing dnaG from the chromosome of SMEG 1 and determination of its DNA sequence. The only departure from the wild-type sequence is a G to C change at base pair position 1488, resulting in a substitution of a proline for an alanine residue at amino acid 496. The same change was also identified in SMEG 15, SMEG 21, and SMEG 23, and these four mutants are probably siblings. We conclude that these mutants are temperature-sensitive due to mutations within the dnaG gene that result in a thermolabile DNA primase.
Organization of the dnaG locus in M. smegmatis.
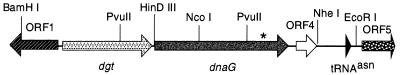
Since dnaG is usually located within the macromolecular synthesis operon of bacteria, we examined the flanking DNA to determine the organization of this locus in M. smegmatis (Fig. 3). This analysis indicated that the M. smegmatis dnaG gene is most likely encoded in an operon with at least one other gene and is organized quite differently from other bacterial macromolecular synthesis operons. The first gene in the operon (ORF2) is located immediately upstream of dnaG, with 53 bp between the stop codon of ORF2 and the start codon of dnaG. Database searching indicated that the putative 428-residue protein product of ORF2 is related to the dGTPase of E. coli (GenBank no. M29995), with an overall identity of 26% and closest similarity at their N termini (Fig. 4). In the Enterobacteriaceae, this enzyme has been shown to be unique in its specificity for dGTP; it catalyzes the reaction dGTP→deoxyguanosine + PPPi. While the physiological role for this enzyme (which is encoded by the dgt gene) is not known, Quirk and Bessman (37) have noted that the enzyme activity appears to be confined to members of the Enterobacteriaceae and did not detect dGTPase activity in extracts of M. smegmatis. Although we cannot rule out the possibility that the M. smegmatis dgt gene is not expressed (or is defective), the observation that M. tuberculosis and M. leprae contain similar dgt genes (see below) suggests that the mycobacteria do encode functional dGTPases. The partial open reading frame (ORF1), upstream of the M. smegmatis dgt gene, is transcribed in the opposite direction to dgt and is of unknown function. Presumably, the signals for expression of both ORF1 and dgt (and probably dnaG) lie within the 59-bp intergenic region between the start codons for ORF1 and dgt.
FIG. 3.
Organization of the dnaG locus in M. smegmatis. The open reading frames identified within a 5.1-kb segment of M. smegmatis DNA are shown as arrows, and the tRNAAsn gene is shown as an arrowhead. ORF1 and ORF5 extend beyond the segment of DNA for which the DNA sequence was determined. The signals for expression of both the divergently transcribed ORF1 and dgt (and dnaG) most likely lie within the 59-bp intergenic region between the start codons for ORF1 and dgt. The dgt gene is located immediately upstream of dnaG. There are 53 bp between the stop codon of ORF2 and the start codon of dnaG. There is a sequence capable of forming a possible RNase III site in the 430-bp intergenic region between ORF4 and the tRNAAsn. Relevant enzyme sites are shown. The position of the mutation conferring the temperature-sensitive phenotype in SMEG 1 is indicated by an asterisk.
ORF4 has no significant similarity to any proteins in the database. ORF4 is predicted to encode a small protein of 102 amino acids and is transcribed in the same direction as dgt and dnaG. However, there is a space of 93 bp between dnaG and ORF4 such that ORF4 could be expressed from signals immediately upstream rather than as part of the putative dgt-dnaG operon (Fig. 3). Downstream of ORF4 there is a tRNAAsn gene, which is of interest in that it is similar to one of the tRNAs encoded by mycobacteriophage L5 (15). There is 68% identity between the L5 tRNAAsn and that of M. smegmatis, indicating that these genes are most likely not derived from each other. There are only five changes between the tRNAAsn from M. smegmatis and those from M. tuberculosis and M. leprae. The only feature of interest identified in the 430-bp region between ORF4 and the tRNAAsn gene is a sequence capable of forming a possible RNase III site just downstream of ORF4.
The last open reading frame identified, the partial ORF5, lies downstream of the tRNAAsn gene (Fig. 3). The search program Beauty (55) revealed that ORF5 has a small region of significant homology to a family of esterase or methyltransferase proteins. One of these proteins, DauP (GenBank no. L35154), is involved in athracycline biosynthesis; another, RdmC (GenBank no. U10405), is thought to be a methyltransferase from Streptomyces purpurascens (12, 34). This sequence similarity (Fig. 4D) seems significant, since a catalytic region, the serine hydrolase active site box G · S · G at positions 100 to 104, is conserved (40). Residues 82 to 122 from ORF5 exhibit 42% identity and 53% similarity to RdmC from S. purpurascens. The alignment of this region (Fig. 4D) also includes sequences from the esterase Est5 (GenBank no. D14529) from Pseudomonas sp. strain KWI-56 (40).
Comparisons of the dnaG loci of M. smegmatis, M. tuberculosis, and M. leprae.
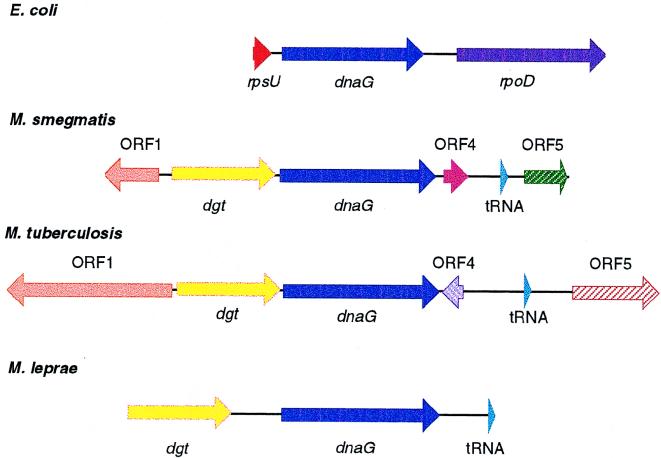
In all prokaryotes that have been studied, the dnaG gene has been found to lie in an operon with rpsU and rpoD, constituting the macromolecular synthesis operon (49, 50). Both B. subtilis and L. monocytogenes have been shown to lack the upstream rpsU, but the dnaG gene is flanked downstream by rpoD (27). The characterization of the dnaG locus in M. smegmatis has revealed a novel arrangement in M. smegmatis as well as in M. tuberculosis (Myc DB cosmid Y98, GenBank no. Z83860) and M. leprae (Myc DB cosmid B1229, GenBank no. L78812) (3) and has allowed comparisons to be made between these loci as shown in Fig. 5. The dnaG locus in M. tuberculosis is located between rrnA and recA near phlC, according to the placement of cosmid Y98 on the integrated map of the M. tuberculosis genome (36). The regions immediately upstream and downstream of the dnaG gene are conserved between these three species and confirm the unique organization of this locus in mycobacteria. The rpsU gene from the macromolecular synthesis operon is replaced by dgt in these three species, whereas the rpoD gene has been replaced by the small ORF4, followed by the downstream tRNAAsn. It is important to note that these coding regions are highly conserved between the genomes of these three species. These observations support the hypothesis that even though these three species are rather distantly related (10), the organization and sequence of the genes determining the central macromolecular processes are highly conserved.
FIG. 5.
Organization of mycobacterial dnaG operons. The organization of the macromolecular synthesis operon of E. coli is shown above the dnaG loci of M. smegmatis, M. tuberculosis, and M. leprae. Comparison of the dnaG loci from M. smegmatis, M. tuberculosis, and M. leprae shows that a region of approximately 4,200 bp between the dgt gene and the tRNAAsn gene is highly conserved among these three mycobacterial species. The dnaG genes are shown in dark blue, and the rpsU gene (shown in red) of the macromolecular synthesis operon is replaced by the putative dgt genes, shown in yellow. The sequence upstream of the putative dgt gene in M. leprae shows similarity to the sequences from M. smegmatis and M. tuberculosis; however, an open reading frame has not been assigned to this region in M. leprae. The partial ORF1 of M. smegmatis and that of M. tuberculosis are shown in orange. ORF4 and ORF5 from M. smegmatis and M. tuberculosis do not have homology and are denoted by different colors. The genes coding for the tRNAAsn are shown in light blue.
The organization of these genes in M. leprae appears to be similar to that in M. smegmatis and M. tuberculosis. However, translation of a full-length DnaG protein requires a frameshift at approximately codon 131, and it is not clear if M. leprae DnaG is expressed via a translational frameshift or if the gene could be nonfunctional; also, the possibility of sequencing errors cannot be ruled out. In addition, we note that the M. leprae DNA sequence upstream of dnaG is closely related to that encoding ORF1 in M. smegmatis and M. tuberculosis, although no open reading frame has been assigned. The tRNAAsn gene is located 456 bp downstream of dnaG in M. leprae (corresponding distances are 837 and 1,054 bp in M. smegmatis and M. tuberculosis, respectively), and no open reading frames have been identified in the intergenic region.
DISCUSSION
The processes of DNA replication and cell division are not well characterized in mycobacteria. Through the characterization of these temperature-sensitive M. smegmatis mutants, the first well-studied DNA replication mutants in mycobacteria, the organization of the dnaG locus in mycobacteria as well as the linkage between cell division and DNA replication in these organisms has been determined. The defect in DNA replication was manifested in the lengthening of the cells at the nonpermissive temperature, indicating that without DNA replication, cell division cannot occur. The linkage of these processes seems obvious and has been characterized in many gram-negative organisms, but this relationship has not been previously observed in mycobacteria.
The abnormal nucleoid appearance indicates that in addition to the DNA replication defect, there may be a problem with chromosome partitioning in SMEG 1, a defect reported for both the parB and dnaG2903 mutants (14, 51, 52). The dnaG2903 mutant was originally isolated by its temperature-sensitive resistance to phenethyl alcohol (14). In E. coli, it is thought that phenethyl alcohol acts at the cell membrane to selectively inhibit the initiation of a new round of DNA replication without interfering with the completion of the ongoing cycle. The dnaG2903 mutant was shown to have a single mutation which was responsible for both the altered membrane property and the temperature-sensitive DNA replication phenotype. Interestingly, in a recent study (7), the E. coli dnaG2903 and parB mutant phenotypes, which are conferred by identical amino acid substitutions in different positions in the extreme carboxy terminus (14), have been shown to be suppressed by era mutants which effectively lower the amount of functional Era, an essential GTPase in E. coli. It is not clear how lowering the amount of wild-type Era or decreasing the functional capacity of Era can overcome the DNA replication-induced cell division defect in these strains, but one possibility is that Era plays a role in cell division (7). It should be noted that the exact role of the primase protein in chromosome partitioning is not known and that there could be a distinct role of primase in this process, or the partitioning defect in the mutants could be due to a secondary effect of perturbation of the DNA replication machinery.
A number of studies have indicated that the carboxy terminus of DnaG is not necessary for the synthesis of primer RNAs (pRNAs). Studies by Sun et al. (46) with proteolytic cleavage products of DnaG showed that the largest fragment, 47 kDa, contains the amino-terminal region of the protein, which exhibits the pRNA synthetic activity of the intact primase in the G4oriC/SSB/primase pRNA synthesis system. However, Sun et al. (46) also showed that the carboxy terminus of primase, a region predicted to stretch from amino acids 398 to 422 to the end of the protein, residue 581 in E. coli, is required for the synthesis of full-length pRNA molecules. These findings suggest that the C terminus of DnaG is needed for processivity or regulation of primase activity. Additional studies by Tougu et al. (47) suggest that a region of the carboxy domain of DnaG interacts with the DnaB helicase at the replication fork. Recent findings by Versalovic and Lupski (51) also indicate that an intact DnaG carboxy terminus is not essential for primase activity; however, it may be necessary for interactions with other DNA replication proteins, such as DnaB, as well as for cell cycle regulatory factors or for the chromosome segregation apparatus (7, 51). The chromosome partitioning defect seen in the parB and dnaG2903 mutants, and now in the M. smegmatis dnaG mutants, further suggests that there may be some essential role provided by primase which links the processes of DNA replication and chromosome partitioning.
The novel organization of this locus in mycobacteria is an indication that there are different regulatory mechanisms for the synthesis of macromolecules in these bacteria. However, the similarity between the dnaG operons of M. smegmatis, M. tuberculosis, and M. leprae suggests that these do not obviously contribute to the differences in the growth rates of these mycobacteria. It seems likely that other bacteria may also depart from the typical macromolecular synthesis operon structure, and we note that Methanococcus jannaschii does not appear to encode a DnaG homolog at all (8). We also note that many of the organisms for which macromolecular synthesis operons have been described are gram-negative enteric organisms and that this conservation might simply reflect their close evolutionary relationship.
It has been hypothesized that mycobacterial genomes are made up of distinct, small regions of DNA conserved between the genomes of mycobacterial species which are flanked by long segments of dissimilar DNAs (36). This mosaic arrangement shows the pressure to conserve small segments of DNA which encode essential functions, while there is a high tolerance for varying arrangements of other coding regions. Through the further characterization of mycobacterial DNA replication mutants, especially strains of the slow growers such as M. bovis BCG, these processes may be directly observed and their regulation may be elucidated.
ACKNOWLEDGMENTS
We thank X. Chang and D. Lever for excellent technical assistance. The mc2155::pYUB415 library and the phasmid pYUB415 were kind gifts from W. R. Jacobs, Jr. Special thanks go to M. Ford for assistance with sequencing. Microscopy and computer assistance were provided by C. Walsh and T. Harper, respectively. Thanks go to G. J. Sarkis for helpful comments on the manuscript. Helpful discussion and insightful comments on the manuscript were provided by J. R. Lupski and R. Hendrix.
This work was supported by grants from the Heiser Foundation to A.G.K., the Howard Hughes Medical Institute Summer Undergraduate Research Fellowship to J.Y.L., and grants from the National Institutes of Health.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baulard A, Jourdan C, Mercenier A, Locht C. Rapid mycobacterial plasmid analysis by electroduction between Mycobacterium spp. and Escherichia coli. Nucleic Acids Res. 1992;20:4105. doi: 10.1093/nar/20.15.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergh S, Cole S T. Myc DB: an integrated mycobacterial database. Mol Microbiol. 1994;12:517–534. doi: 10.1111/j.1365-2958.1994.tb01039.x. [DOI] [PubMed] [Google Scholar]
- 4.Borodovsky M, McIninch J D. Genemark: parallel gene recognition for both DNA strands. Comput Chem. 1993;17:123–133. [Google Scholar]
- 5.Bouche J P, Zechel K, Kornberg A. dnaG gene product, a rifampicin resistant RNA polymerase, initiates the conversion of a single strand coliphage DNA to its duplex replicative form. J Biol Chem. 1975;250:5995–6001. [PubMed] [Google Scholar]
- 6.Brennan P J, Hunter S W, McNeil M, Chatterjee D, Daffe M. Reappraisal of the chemistry of mycobacterial cell walls, with a view to understanding the roles of individual entities in disease processes. In: Ayoub E M, Cassel G H, Bianche W C Jr, Henry T J, editors. Microbial determinants of virulence and host response. Washington, D.C: American Society for Microbiology; 1990. pp. 55–75. [Google Scholar]
- 7.Britton R A, Powell B S, Court D L, Lupski J R. Characterization of mutations affecting the Escherichia coli essential GTPase Era that suppress two temperature-sensitive dnaG alleles. J Bacteriol. 1997;179:4575–4582. doi: 10.1128/jb.179.14.4575-4582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Burton Z F, Gross C A, Watanabe K F, Burgess R R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K-12. Cell. 1983;32:335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- 10.Clark-Curtiss J E. Genome structure of mycobacteria. In: McFadden J, editor. Molecular biology of the mycobacteria. London, England: Surrey University Press; 1990. pp. 77–96. [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickens M L, Ye J, Strohl W R. Analysis of clustered genes encoding both early and late steps in daunomycin biosynthesis by Streptomyces sp. strain C5. J Bacteriol. 1995;177:536–543. doi: 10.1128/jb.177.3.536-543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly-Wu M K, Jacobs W R, Jr, Hatfull G F. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 14.Grompe M, Versalovic J, Koeuth T, Lupski J R. Mutations in the Escherichia coli dnaG gene suggest coupling between DNA replication and chromosome partitioning. J Bacteriol. 1991;173:1268–1278. doi: 10.1128/jb.173.3.1268-1278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatfull G F, Sarkis G J. DNA sequence, structure, and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 16.Ilyina T V, Gorbalenya A E, Koonin E V. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J Mol Evol. 1992;34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs W R, Jr, Bloom B R. Molecular genetic strategies for identifying virulence determinants of Mycobacterium tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: American Society for Microbiology; 1994. pp. 253–268. [Google Scholar]
- 18.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 19.Kinyoun J J. A note on Uhlenhuth method for sputum examination, for tubercle bacilli. Am J Public Health. 1915;5:867–870. doi: 10.2105/ajph.5.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kölbel H K. Electron microscopy. In: Kubica G P, Wayne L G, editors. The mycobacteria: a sourcebook, part A. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 249–250. [Google Scholar]
- 21.Lark K G. Genetic control over the initiation of the synthesis of short deoxyribonucleotide chains in E. coli. Nature (London) New Biol. 1972;240:237–240. doi: 10.1038/newbio240237a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee M H, Pascopella L, Jacobs W R, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy L. Death of Mycobacterium leprae in mice, and the additional effect of dapsone administration. Proc Soc Exp Biol Med. 1970;135:745–749. doi: 10.3181/00379727-135-35134. [DOI] [PubMed] [Google Scholar]
- 24.Levy L. Studies of the mouse foot pad technique for cultivation of Mycobacterium leprae. 3. Doubling time during logarithmic multiplication. Lepr Rev. 1976;47:103–106. doi: 10.5935/0305-7518.19760019. [DOI] [PubMed] [Google Scholar]
- 25.Little J W, Mount D W. The SOS regulatory system of E. coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 26.Lupski J R, Godson G N. The rpsU-dnaG-rpoD macromolecular synthesis operon of E. coli. Cell. 1984;39:251–252. doi: 10.1016/0092-8674(84)90001-1. [DOI] [PubMed] [Google Scholar]
- 27.Lupski J R, Godson G N. DNA→DNA, and DNA→RNA→protein: orchestration by a single complex operon. Bioessays. 1989;10:152–157. doi: 10.1002/bies.950100504. [DOI] [PubMed] [Google Scholar]
- 28.Lupski J R, Ruiz A A, Godson G N. Promotion, termination, and antitermination in the rpsU-dnaG-rpoD macromolecular synthesis operon of E. coli K12. Mol Gen Genet. 1984;195:391–401. doi: 10.1007/BF00341439. [DOI] [PubMed] [Google Scholar]
- 29.Lupski J R, Smiley B L, Blattner F R, Godson G N. Cloning and characterization of the Escherichia coli chromosomal region surrounding the dnaG gene, with a correlated physical and genetic map of dnaG generated via transposon Tn5 mutagenesis. Mol Gen Genet. 1982;185:120–128. doi: 10.1007/BF00333800. [DOI] [PubMed] [Google Scholar]
- 30.Lupski J R, Smiley B L, Godson G N. Regulation of the rpsU-dnaG-rpoD macromolecular synthesis operon and the initiation of DNA replication in Escherichia coli K-12. Mol Gen Genet. 1983;189:48–57. doi: 10.1007/BF00326054. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 32.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1972. [Google Scholar]
- 33.Mustaev A A, Godson G N. Studies of the functional topography of the catalytic center of Escherichia coli primase. J Biol Chem. 1995;270:15711–15718. doi: 10.1074/jbc.270.26.15711. [DOI] [PubMed] [Google Scholar]
- 34.Neimi, J., and P. Mãntsãlã. 1994. GenBank accession no. U 10405.
- 35.Pavelka M S, Jr, Jacobs W R., Jr Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Cole S T. An integrated map of the genome of the tubercle bacillus Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quirk S, Bessman M J. dGTP triphosphohydrolase, a unique enzyme confined to members of the family Enterobacteriaceae. J Bacteriol. 1991;173:6665–6669. doi: 10.1128/jb.173.21.6665-6669.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sarkis G J, Jacobs W R, Jr, Hatfull G F. L5 luciferase reporter mycobacteriophages: a sensitive tool for the detection and assay of live mycobacteria. Mol Microbiol. 1995;15:1055–1067. doi: 10.1111/j.1365-2958.1995.tb02281.x. [DOI] [PubMed] [Google Scholar]
- 40.Shimada Y, Nagao T, Sugihara A, Iizumi T, Yui T, Nakamura K, Fukase T, Tominaga Y. Cloning and sequence analysis of an esterase gene from Pseudomonas sp. KWI-56. Biochim Biophys Acta. 1993;1174:79–92. doi: 10.1016/0167-4781(93)90095-u. [DOI] [PubMed] [Google Scholar]
- 41.Shine J, Dalgarno L. The 3′ terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smiley B L, Lupski J R, Svec P S, McMacken R, Godson G N. Sequences of the Escherichia coli primase gene and regulation of its expression. Proc Natl Acad Sci USA. 1982;79:4550–4554. doi: 10.1073/pnas.79.15.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snapper S B, Lugosi L, Jekkel A, Melton R E, Kieser T, Bloom B R, Jacobs W R., Jr Lysogeny and transformation in mycobacteria: stable expression of foreign genes. Proc Natl Acad Sci USA. 1988;85:6987–6991. doi: 10.1073/pnas.85.18.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 45.Staden R. The current status and portability of our sequence handling software. Nucleic Acids Res. 1986;14:217–231. doi: 10.1093/nar/14.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun W, Tormo J, Steitz T A, Godson G N. Domains of Escherichia coli primase: functional activity of a 47 kDa N-terminal proteolytic fragment. Proc Natl Acad Sci USA. 1994;91:11462–11466. doi: 10.1073/pnas.91.24.11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tougu K, Peng H, Marians K J. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 48.van der Ende A, Baker T A, Ogawa T, Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc Natl Acad Sci USA. 1985;82:3954–3958. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versalovic J, Koeuth T, Britton R, Geszvain K, Lupski J R. Conservation and evolution of the rpsU-dnaG-rpoD macromolecular synthesis operon in bacteria. Mol Microbiol. 1993;8:343–355. doi: 10.1111/j.1365-2958.1993.tb01578.x. [DOI] [PubMed] [Google Scholar]
- 50.Versalovic J, Lupski J R. The Haemophilus influenzae dnaG sequence and conserved bacterial primase motifs. Gene. 1993;136:281–286. doi: 10.1016/0378-1119(93)90480-q. [DOI] [PubMed] [Google Scholar]
- 51.Versalovic J, Lupski J R. Missense mutations in the 3′ end of the Escherichia coli dnaG gene do not abolish primase activity but do confer the chromosome-segregation-defective (Par) phenotype. Microbiology. 1997;143:585–594. doi: 10.1099/00221287-143-2-585. [DOI] [PubMed] [Google Scholar]
- 52.Wada C, Yura T. Phenethyl alcohol resistance in Escherichia coli. A temperature-sensitive mutation (dnaP) affecting DNA replication. Genetics. 1974;77:199–220. doi: 10.1093/genetics/77.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wheeler P R, Ratledge C. Metabolism of Mycobacterium tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: American Society for Microbiology; 1994. pp. 353–385. [Google Scholar]
- 54.Wold M S, McMacken R. Regulation of the expression of the Escherichia coli dnaG gene and amplification of the dnaG primase. Proc Natl Acad Sci USA. 1982;79:4907–4911. doi: 10.1073/pnas.79.16.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worley K C, Wiese B A, Smith R F. Beauty: an enhanced blast-based search tool that integrates multiple biological information resources into sequence similarity search results. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]