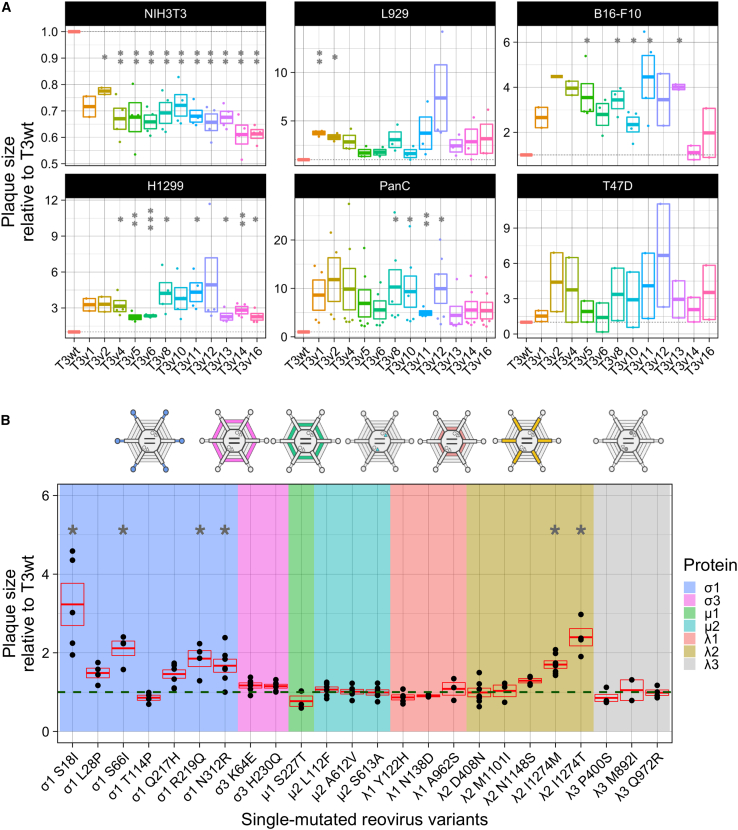
Figure 1.
Isolated mutants selected by directed evolution harbor mutations that increase oncolytic potency evaluated in vitro by plaque size
(A) Plaque size relative to T3wt (set to a value of 1) of original isolated mutants in NIH3T3 (mouse non-transformed fibroblasts), L929 (mouse transformed fibroblasts), B16-F10 (mouse melanoma), H1299 (human lung carcinoma), PanC (human pancreatic carcinoma), and T47D (human breast cancer) cells. Each point represents an independent experiment (n = 2–6). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 relative to T3wt plaque size (p values determined with ANOVA with Tukey test). (B) Plaque size of the 24 reovirus variants harboring single mutations generated by site-directed mutagenesis and reverse genetics. Background color indicates the mutated protein in each virus (see legend). Each point represents an independent experiment performed in duplicate (n = 3–7). The green dashed line corresponds to the plaque size of T3wt and was set to 1. The red box shows mean ± standard error. ∗p < 0.05 relative to T3wt plaque size (p values determined with an ANOVA with Tukey test).