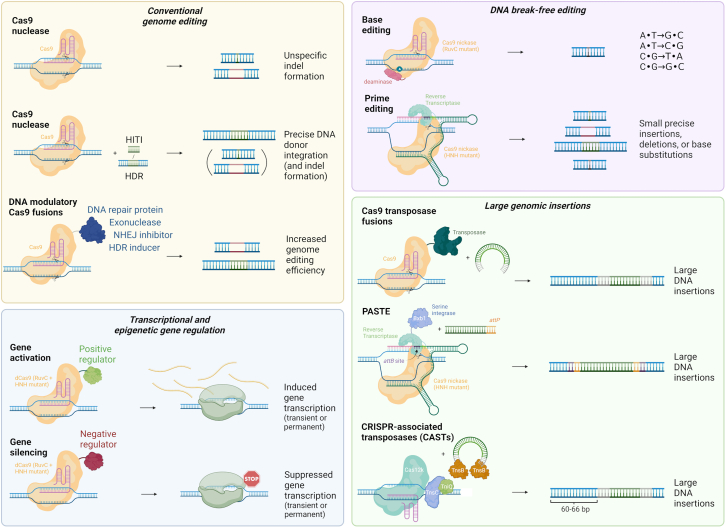
Figure 2.
Gene editing technologies based on CRISPR-Cas systems
The CRISPR-Cas toolbox contains multiple versions of Cas enzymes combined with other proteins to manipulate genomic DNA. For conventional genome editing, Cas9 nucleases are used to create DNA double-strand breaks (DSBs) that facilitate insertions or deletions (indels) of base pairs at the target site introduced by DSB correction via the non-homologous end-joining (NHEJ) pathway, leading to disruption of target DNA sequences. For precise edits, Cas9 nucleases are supplemented with a DNA template for its integration into the desired target locus by either homology-directed repair (HDR) or by homology-independent targeted integration (HITI). These approaches are accompanied by the simultaneous introduction of undesired indels, as such other approaches have fused different DNA modulatory proteins to Cas9 to alter the indel spectrum or to affect the HDR:indel ratio to favor HDR. The DNA break-free base and prime editors (BE and PE, respectively) display high product purity of the editing outcome and highly decrease the risks associated with DNA DSBs, including induction of gross chromosomal aberrations. BE mediates single-base substitutions, while PE can create small precise insertions, deletions, or base substitutions. To induce insertion of large DNA regions, some systems utilize Cas9 fused to transposases, serine integrases (PASTE) or CRISPR-associated transposases (CASTs) to insert large donor DNA templates. PASTE inserts an attB site into the desired genomic location by prime editing, followed by the integration of the donor DNA via the serine integrase (e.g., BxbI) acting on the flanking attP site. The CAST system uses CRISPR-associated transposases to insert transposon DNA engineered to carry the desired cargo. To manipulate the transcriptional status of a gene, a nuclease-deactivated version of Cas9 (dCas9) is employed that maintains the ability to bind a specific DNA target. When fused to transcriptional activators or repressors, target genes can be dialed up or down. By instead using fusion proteins that regulate the epigenetic status of a gene, inherited epigenetic marks can lead to permanent modulation of transcription.