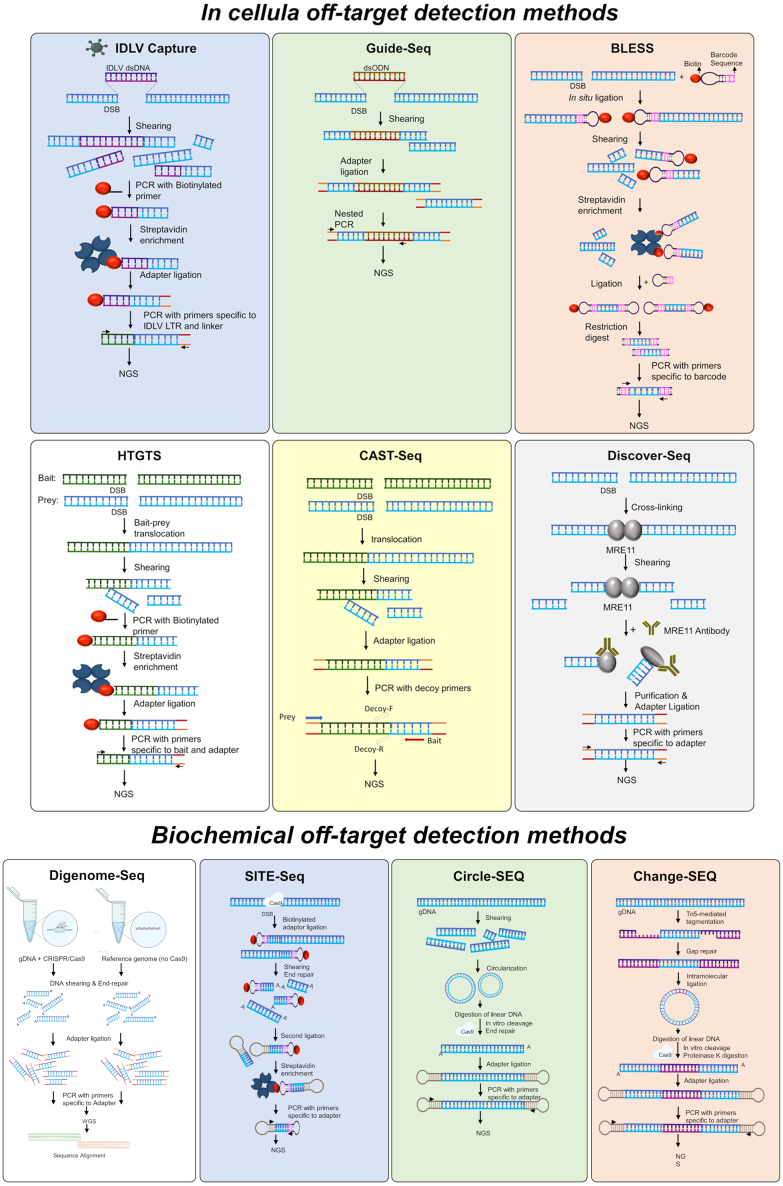
Figure 4.
Sensitive assays and tools for prediction and detection of off-target sites
Schematic overview of in-cellula off-target detection methods. Upper panel, from left to right: integrase-deficient lentivirus capture (IDLV Capture); genome-wide, unbiased identification of DSBs enabled by sequencing (Guide-Seq); direct in situ breaks labeling, enrichment on streptavidin and next-generation sequencing (BLESS). Lower panel, from left to right: high-throughput, genome-wide translocation sequencing (HTGTS); chromosomal aberrations analysis by single targeted linker-mediated PCR sequencing (CAST-seq); discovery of in situ Cas off-targets and verification by sequencing (Discover-seq). DSB, double-stand break; NGS, next-generation sequencing; dsODN, double-strand oligo DNA; LTR, long terminal repeat. Schematic overview of biochemical off-target detection methods, from left to right Digenome-seq, Site-seq, Circle-seq and CHANGE-seq.