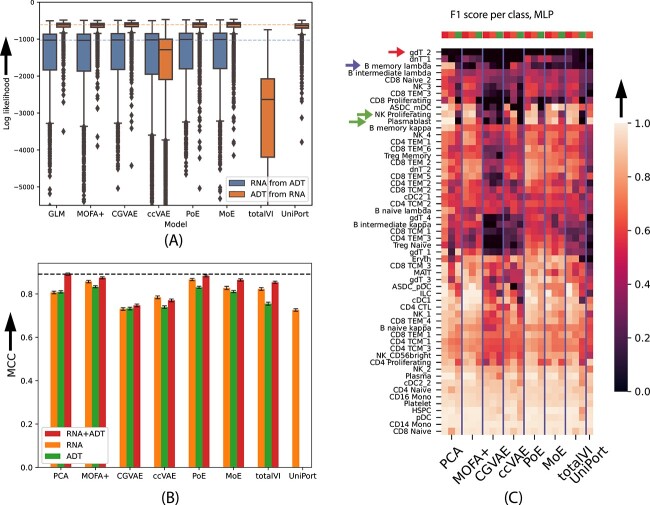
Figure 4.
Evaluation on the CITE-Seq dataset. (A) Missing modality imputation performance for gene expression (RNA) from protein expression (ADT) and vice-versa. Performance is measured as the log-likelihood (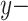 axis, equation 1) of the test samples (cells) given the predictions of each model (
axis, equation 1) of the test samples (cells) given the predictions of each model (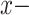 axis) for those data (higher is better). The distribution of the per-cell log-likelihoods is shown. The dashed horizontal lines represent the performance of the baseline GLM. Cells further than 1.5 times the interquartile range from the median are marked as outliers. (B) Cell type classification performance (MCC,
axis) for those data (higher is better). The distribution of the per-cell log-likelihoods is shown. The dashed horizontal lines represent the performance of the baseline GLM. Cells further than 1.5 times the interquartile range from the median are marked as outliers. (B) Cell type classification performance (MCC, 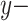 axis, higher is better) achieved by training a multilayer perceptron (MLP) in the joint space of the different models when using: only gene expression (RNA), only protein expression (ADT), and both RNA and ADT data. The error bars denote 95% confidence intervals calculated by bootstrapping the test cells 100 times. (C) Per-class (cell type) performance of the same classifiers as in (B). Brighter colors denote a higher per-class F1 score and therefore better performance. For each model we show three columns (RNA+ADT, RNA only, and ADT only, signified by top row, wherever applicable). Arrows show the cell types highlighted in the results. Note that class CD4+ Tem_4 is not present in the test data and therefore not shown in the per-class evaluations (because its precision and recall is always 0 and the F1 score is thus undefined), but it was taken into account when calculating the MCC in (B).
axis, higher is better) achieved by training a multilayer perceptron (MLP) in the joint space of the different models when using: only gene expression (RNA), only protein expression (ADT), and both RNA and ADT data. The error bars denote 95% confidence intervals calculated by bootstrapping the test cells 100 times. (C) Per-class (cell type) performance of the same classifiers as in (B). Brighter colors denote a higher per-class F1 score and therefore better performance. For each model we show three columns (RNA+ADT, RNA only, and ADT only, signified by top row, wherever applicable). Arrows show the cell types highlighted in the results. Note that class CD4+ Tem_4 is not present in the test data and therefore not shown in the per-class evaluations (because its precision and recall is always 0 and the F1 score is thus undefined), but it was taken into account when calculating the MCC in (B).