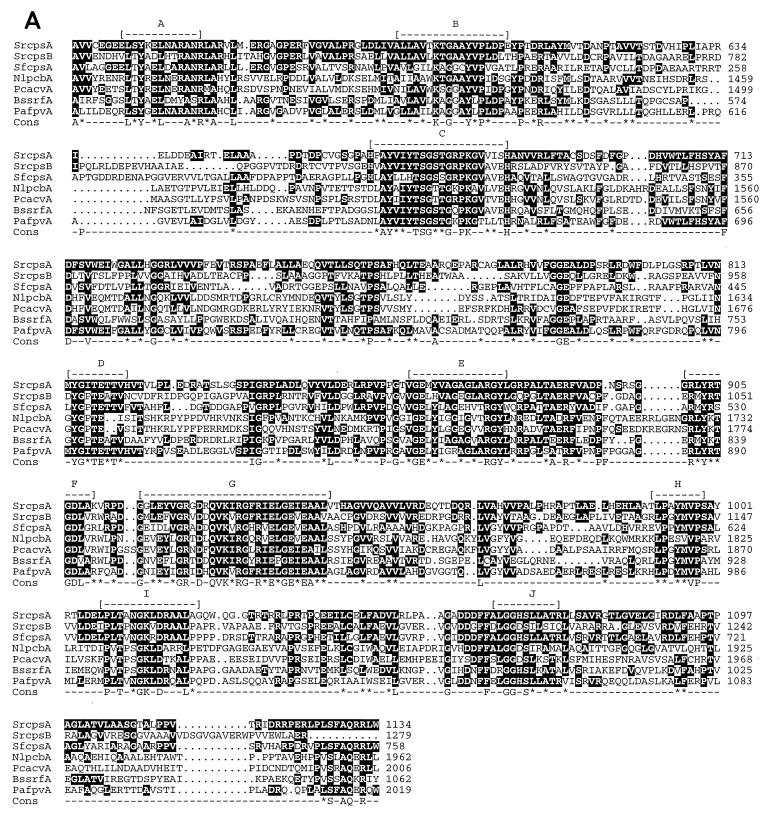
FIG. 3.
Comparison of the deduced amino acid sequences of the peptide synthetase modules. (A) Modules located on the 7.9-kb EcoRI fragment (SrcpsB) and the 5.3-kb ApaI fragment (SrcpsA) (Fig. 2B) are compared to peptide synthetase modules from Streptomyces fradiae A54145 (SfcpsA), N. lactamdurans (NlpcbA), Penicillium chrysogenum (PcacvA), B. subtilis (BssrfA), and Pseudomonas aeruginosa (PafpvA). The conserved motifs (A through J) (33) are shown as dashes. The consensus (Cons) line contains letters for amino acids showing 100% conservation and asterisks for conservative amino acid substitutions. (B) The segment of DNA (Srepim) from the 7.9-kb EcoRI fragment containing epimerase conserved motifs (Fig. 2B) is compared to peptide synthetase segments encoding epimerase activity from Streptomyces pristinaespiralis (Spepim), N. lactamdurans (Nlepim), B. brevis (Bbepim), B. subtilis (Bsepim), and P. crysogenum (Pcepim). The conserved motifs and consensus sequences are depicted as in panel A.