Fig. 2. Analysis of ΔGD and ΔΔGGBE from a set of 27 aptamers:
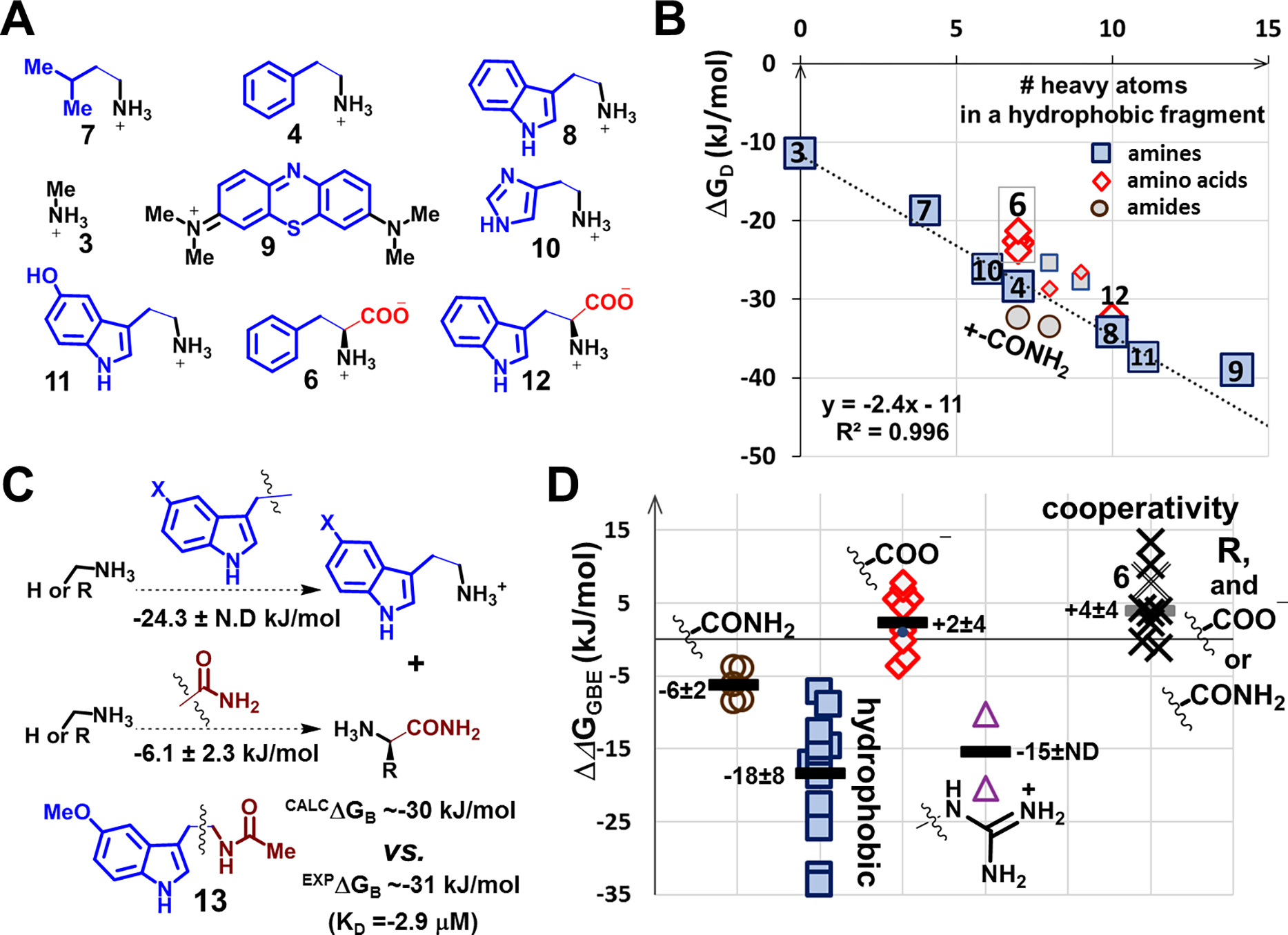
(A) Exemplary targets used to characterize binding optimization to hydrophobic surfaces during selections; hydrophobic/aromatic fragments are shown as brown squares (amines) or green diamonds (amino acids). (B) Regression analysis of target ΔGD vs. # of heavy atoms (other than hydrogen) in aromatic hydrophobic fragments within targets. The regression line including methylamine and two aromatic amines (3, 4, 8) was used to estimate the contributions of the hydrophobic surfaces in two aromatic amino acids (6, 10,) and a non-aromatic hydrophobic amine related to leucine (7). Data for the four aptamers for 6 are shown individually. Methylene blue (9) is the target with the highest affinity for the aptamers isolated directly from N36 libraries. Two amides have ΔGD values above, carboxylates below (diamonds), and histamine (10) and serotonin (11) are on the regression line. Unmarked data points are for tyramine, tyrosine, dopamine and L-DOPA. (C) Additivity of ΔΔGGBE in similar compounds (cf. Fig. S.6): Using the average ΔΔGGBE values of a pair of planar indole-methylene containing molecules and five carboxamides, we estimated ΔGB for the melatonin (13) aptamer. (D) Distributions of ΔΔGGBE contributions of selected functional groups, for carboxylates (open diamonds, we show position of phenylalanine, 6), carboxamides (circles), guanidiniums (triangles), and hydrophobic groups (squares). We show rounded averages (thick lines) and standard deviations in kJ/mol. We also show on the same plot cooperativities (ΔGC) assessed through double functional group replacement cycles (Fig. 1E) for groups added to methylamine together with carboxylates and carboxamides to obtain individual amino acids and their amides (Fig. S41–45). All data points in B-D are results of individual selection experiments, and the uncertainty of this approach can be assessed by four aptamers for phenylalanine, 6, in panel B, which were isolated in four independent selections.
