Fig. 4. Selection of voriconazole aptamers using an analog:
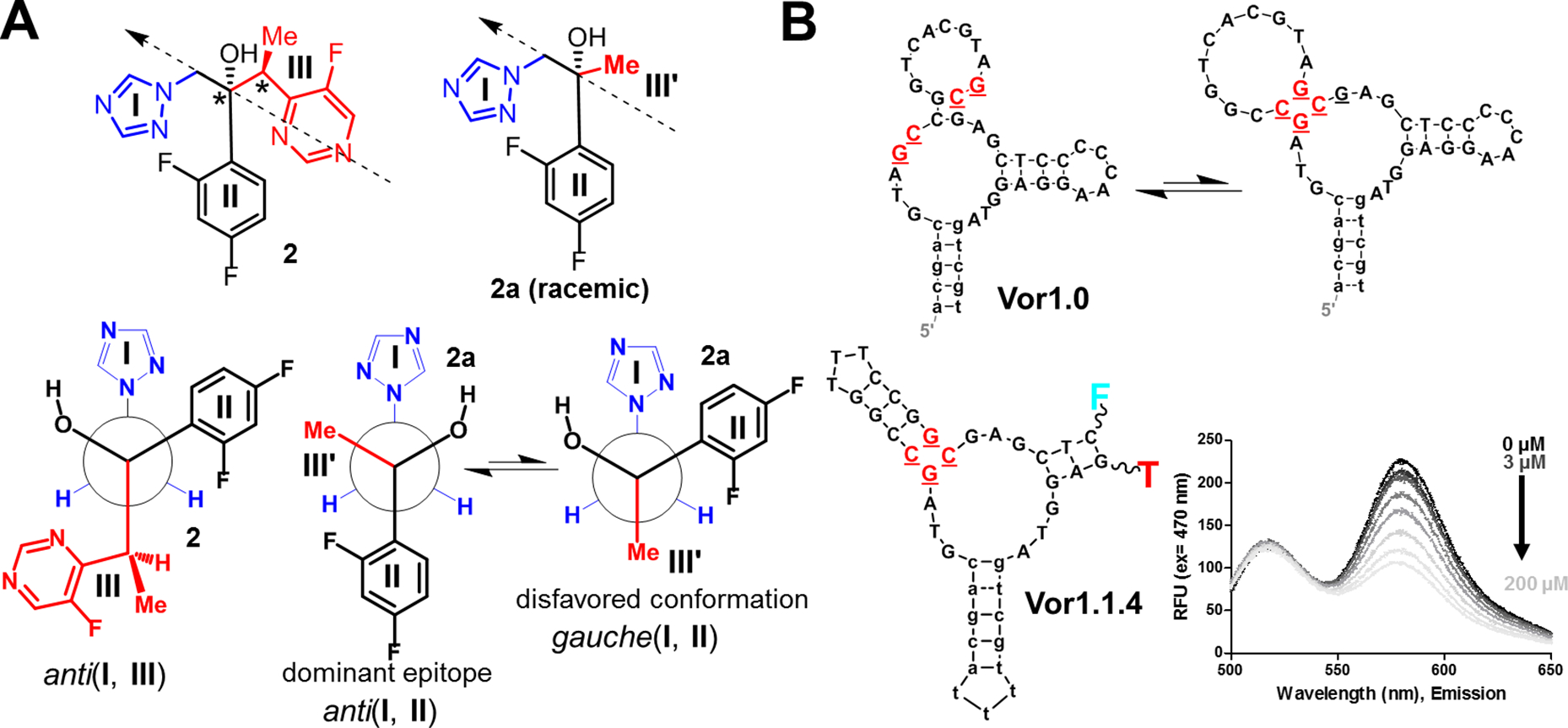
(A) Structure of voriconazole (2) with three fragments (I-III) and its analog 2a, in which fragment III was substituted with a methyl group (III’). The arrows indicate the perspective used to produce the Newman projections below. The anti(I, III) conformation is similar to an observed crystal structure (30). The voriconazole analog 2a simplifies the largest fragment (III) and was designed for reduced complexity and as a more suitable target for selection. Here, the anti (I, II) conformation is likely to be favored and the dominant epitope in selection. (B) The aptamer Vor1.0 was isolated in the selection protocol that used 2 and 2a in parallel. The secondary structure of Vor1.0 is shown as predicted by mFold (top) and as an alternative secondary structure (bottom), which was subsequently confirmed to be the active sensor structure. Structure-switching allows this aptamer to be captured on the column (i.e., the upper structure allows capture) during the initial stages of selection (cf. Fig. S.15). A variant of Vor1.0, Vor1.1.4 (which cannot be captured on the column and, thus, was not isolated during selection) was turned into a quenching-FRET sensor and responded to both 2 and 2.a. Using fluorescence, this sensor detected voriconazole concentrations as low as 3 μM; thus, this oligonucleotide is a candidate for incorporation of electrochemical sensors for in vivo monitoring (12).
