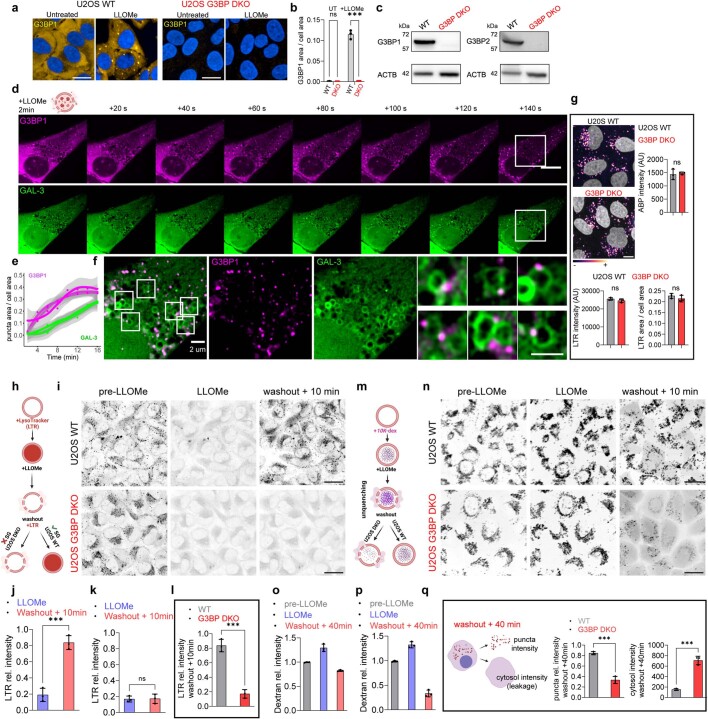
Extended Data Fig. 7. SG dynamics and functional characterisation using U2OS WT and U2OS G3BP double knockout cells.
a-b, Representative images (a) and high content image quantification (b) of U2OS WT and U2OS G3BP DKO cells left untreated or treated with LLOMe (2 mM, 30 min) and stained for G3BP1. Bar plots represent mean values ± SEM of one out of three independent experiments (n = 3), ***P < 0.001, two-way ANOVA, Šídák’s multiple comparisons test. c, immunoblot showing G3BP1 and G3BP2 protein levels in U2OS WT and U2OS G3BP DKO, ACTB was used as a loading control (n = 3). d, Live-cell imaging sequence (20 s time frame) of U2OS WT cells expressing G3BP1-GFP (magenta) and GAL-3 RFP (green) after 2 min of LLOMe treatment (2 mM). e, Shows the quantification of G3BP1 and GAL-3 puncta area over time, indicating that G3BP1-positive signal precedes GAL-3. The curve shows the mean and standard error (shadow areas), n = 3. f, Shows the zoom-in area indicated in (d) (white square), and different areas are further magnified (left panel) highlighting G3BP1+ granules interacting with damaged lysosomes (GAL-3+). g, Representative images of proteolytic activity (magma colormap) in U2OS WT and U2OS G3BP DKO cells incubated with the pan-cathepsin activity-based probe (iABP). Scale bar: 10 μm. Bar plots show iABP intensity quantification (top right), basal LysoTracker puncta intensity (bottom left) and lysosomal area (quantified as LTR area) normalised to cell area (bottom right). h, Scheme illustrating the lysosomal recovery assay using LysoTracker (LTR). i, image sequence of U2OS WT and U2OS G3BP DKO cells incubated with LysoTracker (seen as black puncta) before adding LLOMe (left panel), 2 min after (central panel) and 10 min after washout (left panel). Scale bar: 10 μm. See also Supplementary Video 15. j-k, Show quantification of LTR puncta intensity relative to basal values (pre-LLOMe) for the indicated conditions in U2OS WT (j) and U2OS G3BP DKO (k). n = 3 independent experiments. ***P < 0.001, two-tailed t-test. l, Shows the LTR intensity at 10 min after washout in U2OS WT compared to U2OS G3BP DKO. n = 3 independent experiments. ***P < 0.001, two-tailed t-test. m, Scheme illustrating the lysosomal recovery assay using a dextran-chase assay. Lysosomes loaded with 10 K dextran particles will be damaged using LLOMe and left recover for 40 min after washout (note that, unlike LTR, dextran particles will be unquenched after LLOMe addition). n, Image sequence of U2OS WT and U2OS G3BP DKO cells incubated with 10 K dextran (seen as black puncta) before adding LLOMe (left panel), 2 min after (central panel) and 40 min after washout (left panel). Scale bar: 10 μm. o-p, Show quantification of dextran puncta intensity relative to basal values (pre-LLOMe) for the indicated conditions in U2OS WT (o) and U2OS G3BP DKO (p). n = 3 independent experiments, ***P < 0.001, one-way ANOVA, Dunnett’s multiple comparisons test. q, Shows the dextran puncta intensity and the cytosolic intensity (leakage) at 40 min after washout in U2OS WT compared to U2OS G3BP DKO. Note that since lysosomes do not recover in U2OS G3BP DKO cells, the leakage of dextran particles continues over time resulting in increased fluorescence signal in the cytosol. Data represent the mean ± SEM. n = 3 independent experiments. ***P < 0.001, two-tailed t-test. Scale bar: 10 μm. For gel source data, see Supplementary Fig. 1.