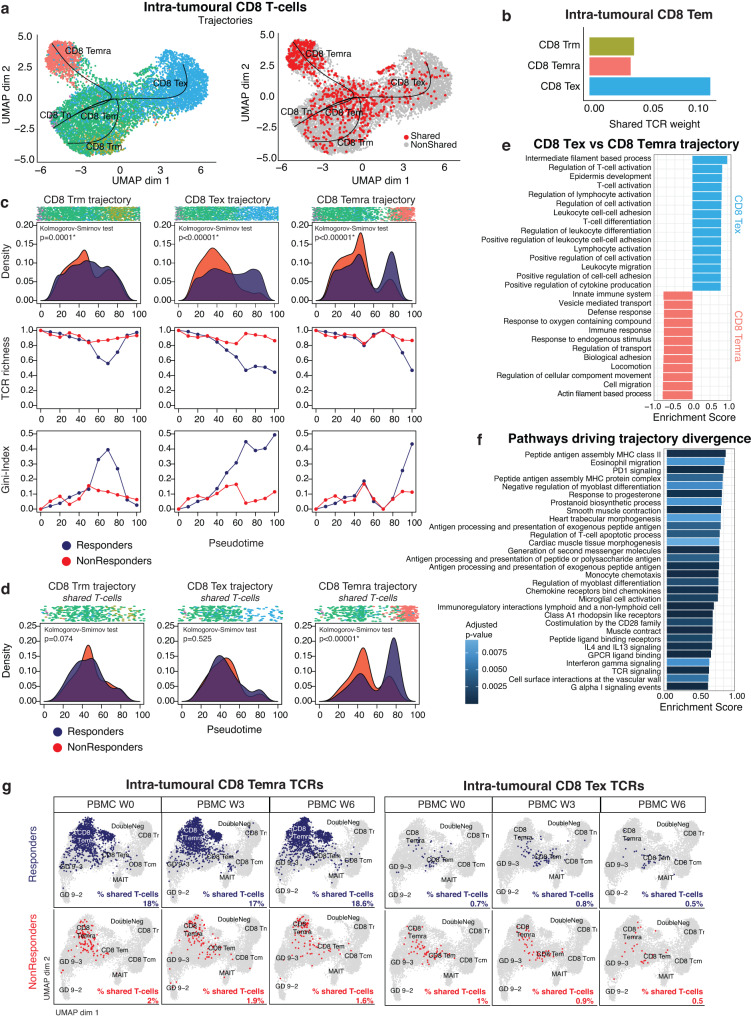
Fig. 4. Interaction with tumour-antigens drives intra-tumoural differentiation towards CD8 TEMRA.
a. UMAP representation of intra-tumoural CD8 T cells (n = 8989). Left: depicting three distinct trajectories: CD8 TRM, CD8 TEX and CD8 TEMRA. Right: depicting shared T cells (i.e. intra-tumoural T cells characterized by a TCR found in PBMC week 0) versus non-shared T cells. b. Bar plot showing the shared TCR weight for CD8 TEM with other CD8 phenotypes in the TME. c. T cell density, TCR richness and Gini-index along each CD8 trajectory in atezo/bev-treated patients (n = 20), stratified for response (12 Resp versus 8 NonResp). The density plots reflect the relative number of T cells separately for Resp versus NonResp along each CD8 trajectory. P-values reflect the difference in distributions, calculated using the two-sided Kolmogorov-Smirnov test. d. Density of shared T cells along each CD8 trajectory in atezo/bev-treated patients (n = 17) stratified for response (10 Resp versus 7 NonResp). The density plots reflect the relative number of shared T cells separately for Resp versus NonResp along each CD8 trajectory. Shared intra-tumoural T cells are characterized by a TCR found in peripheral blood prior to treatment. P-values reflect the difference in distributions, calculated using the two-sided Kolmogorov-Smirnov test. e. Top 15 pathways identified using fGSEA on differentially expressed genes along CD8 TEX versus CD8 TEMRA trajectory (using diffEnd test; TradeSeq35) for the REACTOME and GO: Biological processes gene sets. Significantly enriched pathways (adjusted p-value < 0.01) were identified and ranked based on enrichment score for each gene set separately. Only the top 15 pathways of each gene set, enriched in each trajectory were retained. f. Top pathways identified using fGSEA on differentially expressed genes identified at the point where the three trajectories diverge (using earlyDEG test; TradeSeq35) for the REACTOME and GO: Biological processes gene sets. Adjusted p-value calculated using the Benjamini-Hochberg method. g. UMAP representation of peripheral T cells at week 0 (W0), week 3 (W3) and week 6 (W6) characterized by a TCR shared with CD8 TEMRA (left) and CD8 TEX (right) in the pre-treatment TME, stratified for response to atezo/bev (top: Resp (n = 10), bottom: NonResp (n = 17)). (PBMC, peripheral blood mononuclear cells; TCR, T cell receptor; TME, tumour microenvironment; UMAP, Uniform Manifold Approximation and Projection; Resp, responder; NonResp, non-responder).