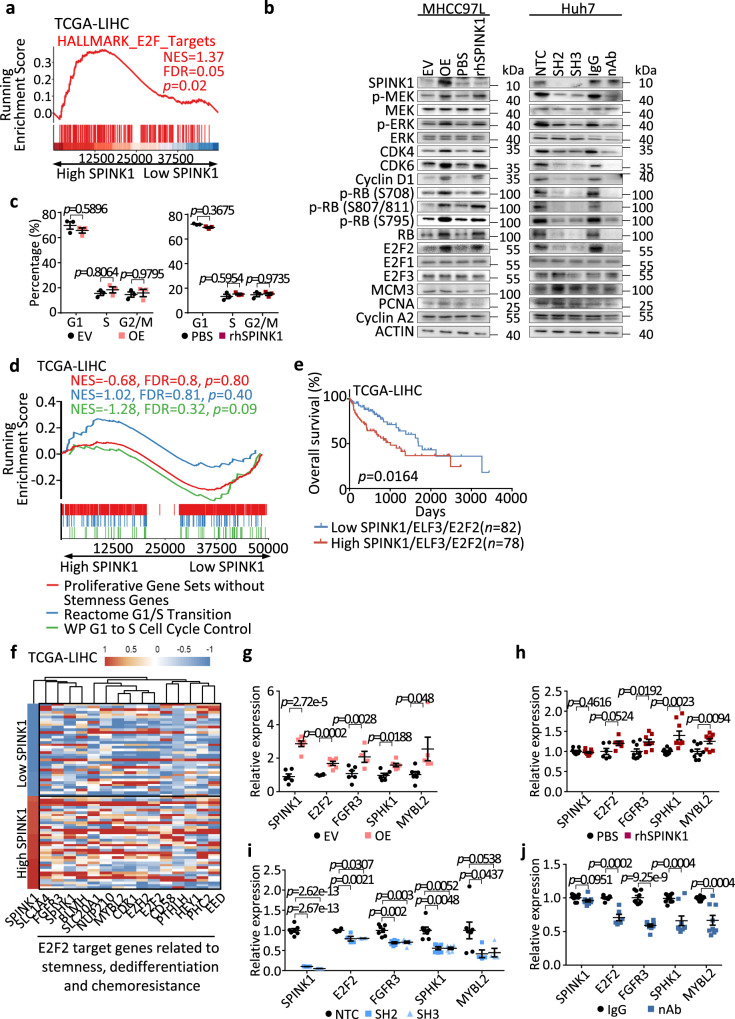
Fig. 8. SPINK1 promotes self-renewal, dedifferentiation, chemoresistance and tumor initiation through a deregulated ERK-CDK4/6-E2F2 regulatory axis.
a E2F targets signature in HALLMARK was enriched in patients with high SPINK1 expression of TCGA-LIHC cohort, via GSEA. b Western blot analysis for expression of SPINK1, phosphorylated and total MEK, phosphorylated and total ERK, CDK4, CDK6, Cyclin D1, phosphorylated and total Rb, E2F2, E2F1, E2F3, MCM3, PCNA and Cyclin A2 in MHCC97L cells with EV versus OE and PBS versus rhSPINK1, and in Huh7 cells with NTC versus SH2 versus SH3 and IgG versus nAb. c Cell cycle analysis of MHCC97L cells (left) with EV or OE, (right) with or without rhSPINK1 by flow cytometry. d GSEA on HCC patients segregated into high/low SPINK1 expression using Reactome G1/S Transition, WP G1 to S Cell Cycle Control, and Proliferative Gene Sets without Stemness Genes signatures in TCGA-LIHC cohort. e Kaplan-Meier curve showing the percentage of overall survival in HCC patients from TCGA-LIHC cohort with high and low SPINK1/ELF3/E2F2 expression. f Heatmap showing the clustering of tumor SPINK1 expression level of TCGA-LIHC cohort with E2F2 target genes related to stemness, dedifferentiation and chemoresistance. g–j qPCR analysis of the expression of SPINK1, and E2F2 target genes, FGFR3, SPHK1 and MYBL2 in MHCC97L cells with g EV versus OE and h PBS versus rhSPINK1, and in Huh7 cells with i NTC versus SH2 and SH3 and j IgG versus SPINK1 nAb. b n = 3 independent experiments; c n = 3 independent experiments; g n = 6 independent experiments; h n = 9 independent experiments; i n = 7 independent experiments; j n = 9 independent experiments. Data were expressed as mean ± s.e.m. Statistical analysis: a, d one-sided Kolmogorov–Smirnov test with adjustments for FDR, c, g–j two-way ANOVA, e log-rank test. Source data are provided as a Source Data file.