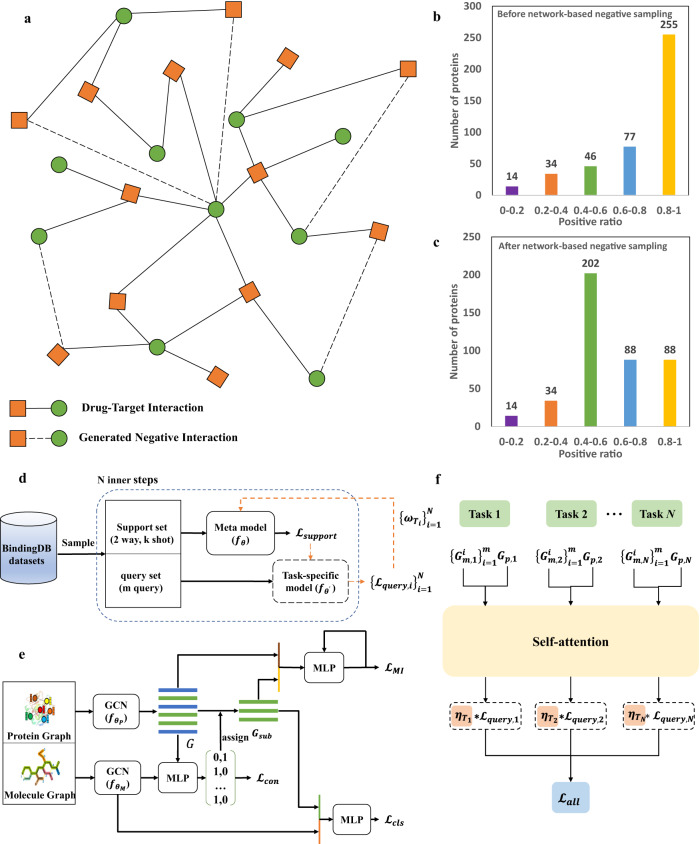
Fig. 1. The framework of ZeroBind.
a Network-based negative sampling strategy. The bipartite network consisting of drugs and protein targets: The square nodes represent the protein nodes and the circle nodes represent the molecule nodes, and there are only edges between different types of nodes, representing the corresponding drug-target interaction. Solid lines represent existing drug-target interactions (DTIs) and dotted lines represent the generated negative interactions with the shortest path distance ≥ 7. b The positive ratio of the training set before the network-based negative sampling strategy. c The positive ratio of the training set after the network-based negative sampling strategy. d Given the support set and query set, is first calculated and utilized to update the base model with parameter to a task-specific model with parameter using the support set of each task, and then the task-specific model calculates the using the query set of the task. After repeating N inner steps, all losses are weighted average by , and gradient descent is further performed to optimize the meta model. e The architecture of the base model in ZeroBind. For each task, the protein graph and the molecule graph are fed into a backbone graph convolutional network (GCN) with parameters and , respectively, to obtain their embeddings. Subsequently, a SIB module is proposed to generate the IB-subgraph of a protein as potential binding pockets in a weakly supervised way. The protein subgraph embedding is concatenated with the molecular embedding and they are fed into a Multilayer Perceptron (MLP) module to identify the interactions. f Task adaptive attention module. It takes the concatenation of the protein embedding and the average of all molecule embeddings in the query set as the task embedding. After using the self-attention layer to compute the weight of each task, denoted as , the overall loss is averaged and incorporated into the meta-training process for updating the model parameters. Source data are provided as a Source Data file.