Abstract
Background
Lung metastasis is the most adverse clinical factor and remains the leading cause of osteosarcoma-related death. Deciphering the mechanisms driving metastatic spread is crucial for finding open therapeutic windows for successful organ-specific interventions that may halt or prevent lung metastasis.
Methods
We employed a mouse premetastatic lung-based multi-omics integrative approach combined with clinical features to uncover the specific changes that precede lung metastasis formation and identify novel molecular targets and biomarker of clinical utility that enable the design of novel therapeutic strategies.
Results
We found that osteosarcoma-bearing mice or those preconditioned with the osteosarcoma cell secretome harbour profound lung structural alterations with airway damage, inflammation, neutrophil infiltration, and extracellular matrix remodelling with increased deposition of fibronectin and collagens by resident stromal activated fibroblasts, favouring the adhesion of disseminated tumour cells. Systemic-induced microenvironmental changes, supported by transcriptomic and histological data, promoted and accelerated lung metastasis formation. Comparative proteome profiling of the cell secretome and mouse plasma identified a large number of proteins involved in extracellular-matrix organization, cell-matrix adhesion, neutrophil degranulation, and cytokine-mediated signalling, consistent with the observed lung microenvironmental changes. Moreover, we identified EFEMP1, an extracellular matrix glycoprotein exclusively secreted by metastatic cells, in the plasma of mice bearing a primary tumour and in biopsy specimens from osteosarcoma patients with poorer overall survival. Depletion of EFEMP1 from the secretome prevents the formation of lung metastasis.
Conclusions
Integration of our data uncovers neutrophil infiltration and the functional contribution of stromal-activated fibroblasts in ECM remodelling for tumour cell attachment as early pro-metastatic events, which may hold therapeutic potential in preventing or slowing the metastatic spread. Moreover, we identified EFEMP1, a secreted glycoprotein, as a metastatic driver and a potential candidate prognostic biomarker for lung metastasis in osteosarcoma patients.
Graphical abstract
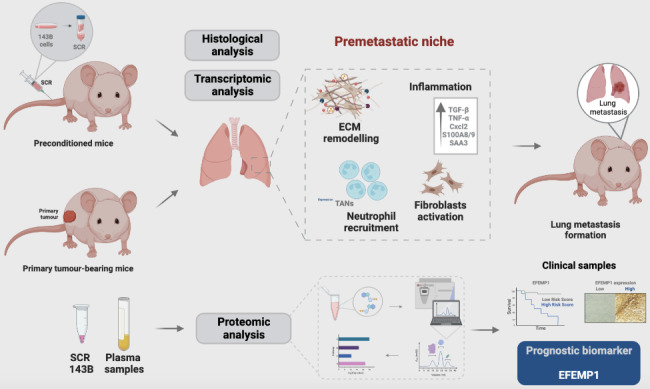
Osteosarcoma-derived secreted factors systemically reprogrammed the lung microenvironment and fostered a growth-permissive niche for incoming disseminated cells to survive and outgrow into overt metastasis.
Daily administration of osteosarcoma cell secretome mimics the systemic release of tumour-secreted factors of a growing tumour in mice during PMN formation;
Transcriptomic and histological analysis of premetastatic lungs revealed inflammatory-induced stromal fibroblast activation, neutrophil infiltration, and ECM remodelling as early onset pro-metastatic events;
Proteome profiling identified EFEMP1, an extracellular secreted glycoprotein, as a potential predictive biomarker for lung metastasis and poor prognosis in osteosarcoma patients.
Osteosarcoma patients with EFEMP1 expressing biopsies have a poorer overall survival.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13046-023-02886-9.
Keywords: Osteosarcoma, Lung Metastasis, Premetastatic niche, Extracellular matrix, Neutrophils, Fibroblasts, EFEMP1
Background
Osteosarcoma (OS) is the most prevalent primary malignant bone tumour that primarily affects children and adolescents [1, 2]. OS has a high propensity to metastasize to the lungs, accounting for 90% of metastatic sites, making it the major cause of morbidity and mortality [3]. The estimated 5-year survival rate of OS patients with localized disease is 70% but drops dramatically to less than 20% in patients who developed metastasis owing to the ineffectiveness of current therapeutic strategies and late diagnosis [4, 5].
It is speculated that over 80% of OS patients have undetectable lung micrometastases at initial diagnosis [6] that eventually progress to death despite receiving aggressive multiagent (neo)adjuvant chemotherapy. Despite intensive efforts to improve the outcome of OS patients, the survival benefits reported in clinical trials evaluating new therapies are quite marginal because of metastatic disease [7]. Mechanistically, the development of distant metastasis is a complex and multi-step process that is distinct from primary tumour formation. The dynamic crosstalk between cancer cells and the local microenvironment is recognized as a critical regulator of tumour progression and metastasis [8–10]. It is now evident that primary tumours prepare in advance and remotely a supportive and receptive microenvironment in specific secondary organs, the so-called premetastatic niche (PMN), for forthcoming tumour cells to adapt and survive [11, 12]. PMN formation encompasses a series of sequential and dynamic events in premetastatic organs primed by tumour-secreted factors and extracellular vesicles (EVs). This process involves a complex interplay of these factors with local stromal cells and tumour-mobilised bone marrow-derived cells (BMDCs) [13, 14]. Vascular leakage, abnormal extracellular matrix (ECM) remodelling, and immunosuppression have been identified as pro-metastatic events [12, 13]. Several studies have already demonstrated these alterations in different tumour types, including melanoma, breast, and colorectal cancers [15–17].
Therapeutic strategies targeting PMN formation at organ-specific sites offer an opportunity to prevent or suppress metastasis formation and are currently a hot topic in cancer research [18, 19]. The identification of potential druggable targets requires a tumour-specific understanding of the cellular and molecular mechanisms involved in establishing organ-specific PMNs. Despite advances in this topic, sarcomas have been less studied, and the mechanisms underlying the development of organotropic lung metastasis are not clearly understood [5, 20].
The cancer secretome is a reservoir of potential biomarkers and signalling biomolecules relevant to tumour progression and metastasis [21, 22]. This class of proteins has been studied in the search for cancer biomarkers and to understand their mechanistic role in tumour progression [23, 24]. Jerez and colleagues [25] conducted a proteomic analysis of the secretome of human OS cells and identified specific proteins in both exosomes and soluble fractions, involved in biological functions (angiogenesis, cellular adhesion, and migration) related to tumour progression and metastasis. Recently, Mazumdar et al. [26] explored the functional role of OS-derived EVs in driving lung metastatic colonization. Despite having observed preferential accumulation of EVs in the lungs, the induced stromal changes did not result in an increased tumour burden, suggesting that EVs dictate organotropism but additional tumour-secreted factors are required to establish a functional PMN [26].
In this study, we evaluated how primary OS systemically reprograms the lung microenvironment to establish a permissive pro-metastatic niche for subsequent metastasis formation using two murine models and a multi-omics approach. Transcriptomic data and tissue analysis uncovered ECM remodelling with deposition of cell adhesion-promoting proteins by stromal-activated fibroblasts, and neutrophil recruitment as early pro-metastatic events guiding lung metastasis. Furthermore, we identified EFEMP1, a glycoprotein secreted by tumour cells, as a key player in the development of lung metastasis and a potential prognostic biomarker in OS patients.
Methods
Cell culture and Infection with a lentivirus encoding luciferase
The human OS cell line 143B was purchased from the American Type Culture Collection (ATCC, Manassas, USA) and cultured in Eagle’s minimum essential medium (EMEM, Sigma Aldrich, UK), supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco, Paisley, UK), 0.015 mg/ml 5-bromo-2’-deoxyuridine (BrdU, Sigma Aldrich, St. Louis, MO, USA), 1.0 mM sodium pyruvate (Gibco, Grand Island, USA), and 1% antibiotic-antimycotic (Gibco, Grand Island, USA). The cells were maintained under standard adherent conditions in a humidified incubator with 5% CO2 at 37 °C. Cells were authenticated by immunohistochemistry with antibodies against vimentin (Invitrogen, Thermo Fischer Scientific, Netherlands) and Ki-67 (clone MIB-1; Dako) and haematoxylin and eosin (H&E). The 143B cells were stably transduced with a lentivirus encoding Luciferase as described elsewhere [27].
For secretome (SCR) preparation, 143B cells (25 × 103 cells/cm2) were cultured in an exosome-free culture medium for 24 h. The exosome-depleted medium was prepared by ultracentrifugation of 50% FBS in the appropriate cell culture medium (120 000 g, 16 h). The supernatant was collected after 24 h of incubation, centrifuged to remove cell debris and concentrated with Amicon Ultra-15 Centrifugal Filters (10 kDa molecular weight cut-off, Merck Millipore Ltd, Carrigtwohill, Ireland). Aliquots were stored at -80ºC until usage.
EFEMP1 knockdown
Small interfering RNA (siRNA) knockdown of EFEMP1 in 143B cells was performed using the siGENOME human EFEMP1 siRNA SMARTpool (M-011855-01-0010). At 50% confluency cells were transfected with 20nM siRNA and a nontargeting (NT) control sequence (D-001810-10), both from Dharmacon Inc (Lafayette, USA), using Lipofectamine™ 3000 (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer’s recommendations. After 48 h, the culture medium was replaced by an exosome-free medium, and the cells were maintained in culture for another 24 h afterwards the secretome was collected. The knockdown efficiency was evaluated by western blot and ELISA.
Animal studies
Athymic Swiss nude (Foxn1nu/nu) mice, male or female, (8–12 weeks old, 20–30 g) were purchased from the Institute for Clinical and Biomedical Research (iCBR, Coimbra, Portugal) of the Faculty of Medicine of the University of Coimbra and housed under pathogen-free conditions in individually ventilated cages, with controlled temperature/humidity (22 °C/55%) environment on a 12 h light-dark cycle and with food and water ad libitum.
Premetastatic niche formation
Animals were randomized into two groups. A group was injected subcutaneously into the lower flank with 5 × 105 of 143B-Luc+/100 µL PBS for primary tumour formation. The tumour growth was monitored every two days in two dimensions using a digital calliper, and mice were sacrificed when the tumour reached 50–60 mm3 in volume. Tumour volumes were calculated using the modified ellipsoid formula V = A × B2/3 (A length; B width).
The second group received a daily intraperitoneal (i.p.) injection of 25 µL of the 143B-derived SCR or the vehicle for 1 week, following which they were sacrificed. Animals were euthanized by cervical dislocation, and peripheral blood and lung tissue were harvested for further analysis. Peripheral blood samples were collected to K3EDTA tubes (Greiner bio-one, Kremsmunster, Austria) from anaesthetized animals via cardiac puncture before euthanasia. Plasma was separated by centrifugation at 3000 rpm for 10 min at 4ºC, and aliquots were stored at -80ºC until usage.
Mouse models of Lung Metastasis
For experimental lung metastasis formation, animals were injected intravenously (i.v.) into the lateral tail vein with 1.5 × 106 of 143B-Luc+ cells/100 µL PBS. Another set of animals received daily intraperitoneal injections of 25 µL of the SCR for 1 week prior to the i.v. injection of 143B-Luc+ (1.5 × 106 cells/100 µL PBS). A separate group received the SCR of EFEMP1-knockdown 143B cells, followed by tail vein injection of the 143B-Luc+ cells, as described above.
For spontaneous lung metastasis formation, animals were injected subcutaneously in the lower flank with 5 × 105 of 143B-Luc+ cells/100 µL PBS. After reaching a maximum volume of 60–100 mm3, subcutaneous tumours were surgically excised and the skin incision was sutured with the animals under anaesthesia (10 mg/kg; Rompun 2%, Kiel, Germany).
Animals were monitored weekly for lung metastasis formation during a maximum of 60 days by bioluminescence imaging (BLI) on an IVIS Lumina XR (Caliper Life Sciences Inc., PerkinElmer, Massachusetts, USA). Images were acquired after i.p. injection of D-Luciferin (150 mg/kg), with the animals anaesthetized with 2.5% of isoflurane (Virbac, Carros, France) in 100% O2. Bioluminescent images were analysed using the Living Image software version 4.10 (Xenogen, Alameda, California). A region of interest was drawn around the lesions for the quantification of the bioluminescent signal. Values are expressed photons/sec/cm2/sr. At the end of the study, animals were euthanized by cervical dislocation and peripheral blood and metastatic lungs were collected for histopathological analysis.
Clinical samples
Paraffin-embedded biopsy specimens were obtained retrospectively from 29 patients diagnosed with OS. At the time of diagnosis, all patients had strictly localized disease and were naïve to any chemotherapy. Of the 29 patients, 26 were classified as having conventional high-grade OS while the remaining 3 were classified as low-grade. Except for those with low-grade, all patients underwent chemotherapy prior to surgery, and all received adjuvant chemotherapy. The follow-up period ranged from 1 year to at least 10 years, or until death.
The specific characteristics of the patients are summarized in Table 1.
Table 2.
Primer sequences used in qRT-PCR
| Gene | Forward (5’ – 3’) | Reverse (5’ – 3’) |
|---|---|---|
| CCL2 | CAAGATGATCCCAATGAGTAG | TTGGTGACAAAAACTACAGC |
| CXCL2 | GGGTTGACTTCAAGAACATC | CCTTGCCTTTGTTCAGTATC |
| CXCR2 | CTACTGCAGGATTAAGTTTACC | GACGTATATTACAACCACAGC |
| FN1 | CCTATAGGATTGGAGACACG | GTTGGTAAATAGCTGTTCGG |
| GAPDH | GCCTTCCGTGGTCCTACC | GCCTGCTTCACCACCTTC |
| HPRT1 | GTTGAAGATATAATTGACACT | GGCATATCCAACAACAAA |
| IL-6 | TTCCATCCAGTTGCCTTC | TTCTCATTTCCACGATTTCC |
| IL-1β | TCTATACCTGTCCTGTGTAATG | GCTTGTGCTCTGCTTGTG |
| PRG4 | GATAATGCTATTCCAGGCAC | CATCCAGAAATAATGACCTCG |
| S100A8 | ATACAAGGAAATCACCATGC | ATATTCTGCACAAACTGAGG |
| S100A9 | CTTTAGCCTTGAGCAAGAAG | TCCTTCCTAGAGTATTGATGG |
| TGF-β | ATGGTGGACCGCAACAAC | TTGCTATATTTCTGGTAGAGTTCC |
| TNF-α | CAAGGGACTAGACAGGAG | TGCCTCTTCTGCCAGTTC |
Table 1.
Clinicopathological features of osteosarcoma patients
| Number of patients | 29 |
|---|---|
| Age | |
| Mean (range) | 33.6 (15–78) |
| Variable | Number (%) |
| Gender | |
| Male | 15 (52) |
| Female | 14 (48) |
| Primary tumour location | |
| Femur | 16 (55) |
| Tibia | 5 (17) |
| Humerus | 2 (7) |
| Fibula | 2 (7) |
| Others | 4 (14) |
| Tumour grade | |
| High grade | 26 (90) |
| Low grade | 3 (10) |
| Metastasis | 12 (41) |
| Disease-free interval (DFI) | Months |
| Mean (range) | 41.4 (6-159) |
Scanning electron microscopy (SEM)
Small fragments of resected lung tissues and decellularized matrices were fixed with 2% glutaraldehyde and examined in a scanning electron microscope (Flex SEM 1000, HITACHI), under variable pressure scanning using an accelerating voltage of 5–10 kV.
Transmission electron microscopy (TEM)
Resected lung tissues were sectioned into small fragments (1 mm3) and fixed for 2 h in 2.5% glutaraldehyde buffered with 0.1 M cacodylate buffer (pH 7.4), followed by a post-fixation in 1% osmium tetroxide (OsO4) for 1.5 h. After washing, samples were incubated with 1% aqueous uranyl acetate for 1 h, for contrast enhancement. Samples were then dehydrated in a graded acetone series (30–100%) followed by resin embedding using an epoxy embedding kit (Fluka Analytical, Sigma Aldrich, Germany). Ultra-thin sections were obtained with a Leica EM UC6 (Leica Co; Austria) ultramicrotome, mounted on copper grids and stained with lead citrate 0.2% for 7 min. Images were acquired on a Tecnai G2 Spirit Bio Twin electron microscopy at 100 kV (FEI) and AnalySIS 3.2 software.
Histopathological analysis and immunostaining
For murine models, lung tissues and primary tumours were fixed in 4% paraformaldehyde (PFA) and processed for paraffin embedding. Sections of 4 μm were stained with haematoxylin and eosin (H&E, Sigma-Aldrich, St. Louis, MO, EUA) or antibodies against fibronectin (ab2413, Abcam, USA), alpha-smooth muscle actin (α-SMA, ABT 1487, Millipore, Darmstadt, Germany) and vimentin (V9; Ventana, Arizona, EUA). Antigen retrieval was performed by immersing slides in EDTA-Tris buffer (pH 8) for 8 min at 95 ºC and then blocked with a buffered hyper protein solution for 4 min to avoid nonspecific bonds. Immunostaining was performed using a Ventana Marker Platform Benchmark Ultra IHC/ISH with the resource of a multimeric indirect free biotin detection system - Optiview DAB IHC Detection Kit (Ventana Medical Systems, Arizona, EUA), according to the manufacturer instructions. A Gordon’s and Sweet silver staining was performed for the detection of reticulin fibers. Slides were observed under a light microscope Nikon Eclipse 50 I and images were captured with a Nikon-Digital Sight DS-Fi1 camera. Architectural changes were evaluated, and inter-alveolar septal thickness was measured in randomly selected H&E-stained sections.
Clinical samples were formalin-fixed for 24 h and then decalcified with 10% nitric acid for 1–5 days based on tissue hardness, with daily integrity checks. The samples were paraffin-embedded and sectioned into 4 μm tissue slices for immunostaining with anti-EFEMP1/fibulin 3 monoclonal antibody (1:500, ab256457, Abcam). Hematoxylin-eosin staining was performed using standard methods as a counterstain. Immunohistochemistry positivity was considered when at least 1% of the viable neoplastic cells showed cytoplasmic and/or membrane expression of any intensity (weak, moderate or strong). The sections were assessed by an experienced pathologist.
Primary lung fibroblasts isolation and culture
Lungs were harvested, washed in PBS, minced with scissors, and enzymatically digested with 0.1% collagenase A (Roche, Mannheim, Germany) and dispase II (2.4 U/mL; Gibco, Japan) for 90 min at 37 ºC, and then filtered through a 70 μm filter strainer and washed with a physiological saline solution containing 0.05 M EDTA. The obtained cell suspension was plated into 1% gelatin pre-coated dishes and maintained in RPMI 1640 (Sigma-Aldrich, UK) supplemented with 15% FBS and 1% antibiotic-antimycotic at 37 ºC under 5% CO2. All experiments were performed with early passages fibroblasts (P2-P4).
Decellularization
Lung fragments
Small lung fragments (2 × 2 mm) were incubated in hypotonic buffer (10 mM Tris HCl/0.1% EDTA, pH 7.8) for 18 h at room temperature, and then washed in PBS (3x, 1 h) and immersed in a detergent solution (0.2% SDS/10 mM Tris HCl, pH 7.8) for 24 h at room temperature. Fragments were washed in hypotonic buffer (3x, 20 min) and incubated in DNAse solution (50 U/ml DNAse/ 10mMTris HCl, pH 7.8) for 3 h at 37 °C. Decellularized matrices were washed in PBS (2x, 20 min) to remove the residual detergent and DNAse solution and were maintained under sterile conditions at 4ºC in PBS until use. All steps were performed under agitation at 165 rpm.
Fibroblasts
Fibroblast cell sheets were incubated with buffer I (1 M NaCl, 5 mM EDTA, 10 mM Tris/HCl pH 7.4) for 1 h at room temperature, washed with PBS (3x, 10 min) and then exposed to buffer II (0.5% [w/v] SDS, 25 mM EDTA, 10 mM Tris/HCl pH 7.4) for 30 min with agitation.
Immunofluorescence
Primary fibroblasts were fixed with 4% paraformaldehyde (PFA, Sigma-Aldrich, St. Louis, MO, USA) for 20 min, permeabilized with 0.2% TritonX-100 (Sigma-Aldrich, St. Louis, MO, USA) for 10 min, and blocked with 3% bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO, USA) for 1 h at room temperature. Afterwards, the cells were incubated with the primary antibodies against α-SMA (1:200, ABT 1487; Millipore, Darmstadt, Germany), fibroblast activation protein (FAP) (1:150, PA5-99313, Thermo Fisher Scientific, USA), vimentin (1:200, SP20, Thermo Fisher Scientific, USA) and fibronectin (1:200, ab2413, Abcam, Cambridge, UK) overnight at 4 ºC. The fibroblasts were then incubated with secondary antibody Alexa Fluor 568 or 488 (1:200, Invitrogen, USA) for 1 h at room temperature in the dark, and nuclei were counter with 2 mg/mL Hoechst 33,342 (Sigma-Aldrich, USA).
For immunofluorescence staining with Alexa Fluor 555 phalloidin, cells were fixed with PBS containing 4% sucrose and 4% paraformaldehyde for 10 min, permeabilized with 1% TritonX-100 for 10 min and blocked with 1% BSA for 45 min at room temperature , and then incubated with Alexa Fluor 555 phalloidin (1:40, Abcam, Cambridge, MA, USA) for 30 min at RT. Nuclei were stained with 2 mg/mL Hoechst 33,342 (Sigma-Aldrich, Buchs, Switzerland). The chamber slides were mounted in the Vectashield Mounting Medium (Vectashield, Vector Laboratories, United States).
Decellularized lung fragments and fibroblast cell sheets were processed for immunofluorescence as described above and stained with the primary antibodies against fibronectin (1:200) and collagen IV (1:200 ab19808; Abcam, Cambridge, UK) and then incubated with secondary antibody Alexa Fluor 488 (1:200, Invitrogen, USA). Images were captured in Carl Zeiss Axio Observer Z1 inverted microscope (Carl Zeiss, Thornwood, NY) and processed using the ImageJ 1.52p software (National Institutes of Health, USA).
Cell migration
Cell migration was analysed using the wound-healing assay. Confluent monolayers of fibroblasts were manually scratched with a 200 µL sterile pipette tip. Photographs were taken immediately after scratching (baseline) and at 6 and 24 h in a Carl Zeiss Axio Observer Z1 inverted microscope (Carl Zeiss, Thornwood, NY). The wound closure at each time-point was quantified using ImageJ 1.52p software (National Institutes of Health, USA) and normalized to the baseline.
Lung transcriptomic analysis
Total RNA was extracted from lung tissues using TRIzol reagent (Ambion by Life Technologies, USA) according to the manufacturer’s instructions. RNA was eluted in 30–40 µL RNase-free water. The concentration and quality of extracted RNA were determined with a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, USA). RNA was stored at 80 ºC until use. Single-end sequencing was performed using the library prep kit TruSeq and the sequencing kit NovaSeq6000 SP Flowcell 100 cycles (Illumina, Inc.) for Mus musculus (Ensembl.GRCm38.82). The transcriptomic sequencing of RNA was performed on the Illumina NovaSeq 6000 instrument, at the VIB Nucleomics Core (www.nucleomics.be). The reads pre-processing was performed by VIB Nucleomics Core and involved: quality trimming (FastX 0.0.14, HannonLab), adapter trimming (cutadapt 1.15) [28], quality filtering (FastX 0.0.14 and ShortRead 1.40.0) [29] and removal of contaminants (bowtie 2.3.3.1). The pre-processed reads were then aligned to the reference genome of Mus_musculus.Ensembl.GRCm.38.82 using STAR 2.5.2b [30] and SAMtools 1.5 [31] Bioconductor package. The expression levels of the read overlapping genes were computed using the EDASeq package [32] for the within-sample and between samples normalizations. The differentially expressed genes were estimated by fitting a negative binomial generalized linear model using the edgeR 3.24.3 package [33]. The resultant p-value was corrected for multiple tests with Benjamini-Hochberg to control the false discovery rate (FDR).
Unless stated otherwise, the following bioinformatics analyses were conducted using the R programming language (version 4.1.1) in the RStudio integrated development environment. The volcano plots were achieved using the ggplot2 [34] (version 3.3.5) and ggrepel (version 0.9.1) packages. The Venn diagrams were generated with the webtool available at bioinformatics.psb.ugent.be/webtools/Venn/. The clusterProfiler [35] version 4.0.5) package was used to perform the gene set enrichment analyses (GSEA) for the Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Genes showing a p-value higher than 0.05 were excluded from the gene set. Heat maps were made with the ComplexHeatmap [36] (version 2.8.0) package. Protein-protein interaction (PPI) network of DEGs was visualized using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/). Graphics were refined and figures were assembled in Adobe Illustrator 2019 (version 23.1.1).
Flow cytometry
Fresh lung tissue was minced with fine scissors and subjected to mechanical dissociation (GentleMACS, Miltenyi Biotech, Germany). The tissue homogenates were filtered gently through a 40 µm cell strainer and centrifuged at 600 g for 15 min at RT. The isolated single-cell suspension and peripheral leukocytes were fluorescently stained with anti-mouse CD11b-FITC (1:200, BioLegend, 101205) and Ly6G-PE (1:80, BioLegend, 127607), CD68-APC (1:300, BioLegend, 137007), CD19-PerCP (1:200, BioLegend, 115531) and CD45-PB (1:200, BioLegend, 103125). FACS Lysing solution (BD Sciences) was added to the samples for lysing red blood cells. Data were collected on a FACS Canto™ II (BD Biosciences, USA) and analysed using Infinicyt V.1.8 software (Cytognos, Salamanca, Spain).
RNA extraction and qRT-PCR
Total RNA from lung tissue was extracted using TRIzol reagent (Ambion by Life Technologies, USA) according to the manufacturer’s instructions. The concentration and quality of extracted RNA were determined using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, USA). First-strand complementary deoxyribonucleic acid (cDNA) was synthesized from 2 µg of total RNA using the NZY first-strand cDNA synthesis kit. Quantitative real-time PCR analysis was performed using a Xpert SYBR Green MasterMix (GRISP, Porto, Portugal) on a CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories, Inc). Gene expression was normalized to the housekeeping genes glyceraldehyde 3-phosphate dehydrogenase (GADPH) and hypoxanthine-guanine phosphoribosyltransferase 1 (HPRT-1) ranked as stably expressed by the RefFinder algorithm (https://www.heartcure.com.au/reffinder/). The relative expression of target genes was calculated based on the 2−∆∆Ct method, where ΔΔCt=(Ct target−Ct housekeeping)Sample−(Ct target−Ct housekeeping)Control. Primer sequences (Eurofins Genomics, Lisboa, Portugal) are listed in Table 2. A Primer-BLAST search was performed to evaluate primer specificity and self-complementarity values of already validated and published primer sequences.
Western blot
Total protein extracts were prepared using standard lysis buffer, separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). Membranes were blocked with 5% of bovine serum albumin in Tris-buffered saline-tween 20 (TBS-T) and were incubated with appropriate primary antibodies fibronectin (1:500; ab2413; Abcam, Cambridge, UK), collagen IV (1:500; ab19808; Abcam, Cambridge, UK), EFEMP1 (1:1000, ab256457, Abcam), ITG β1 (1:1000, D6S1W, Cell Signalling), ITGα6 (1:1000, ab181551, Abcam) and CD44/HCAM (1:200,sc-7297, Santa Cruz Biotechnology), overnight at 4 °C. Afterward, the membranes were washed in 0.1% TBS-Tween and incubated with the secondary antibody (1:10000) for 1 h at RT and revealed by chemiluminescence (Clarity ECL western blotting substrates, BioRad) using ImageQuant LAS 500 chemiluminescence CCD camera (GE Healthcare Bioscience AB). Images were analysed using ImageJ software (National Institutes of Health).
Adhesion assays
The 96-well plates were coated with fibronectin or collagen at concentrations ranging from 1 to 10 µg/mL for 2 h. The 143B-luc+ cells were seeded onto the substrates and allowed to adhere for 10 min at 37 °C. To remove the non-adherent cells, the wells were washed 3 times with PBS containing 1 M CaCl2.H2O and MgCl2.6H2O, and then 100 µL of EMEM culture medium containing 0.3 mg/mL of D-Luciferin was added to each well. The plate was read in the IVIS optical imaging system, and the bioluminescent signal was quantified using Living Image Software 4.10 (Xenogen, Alameda, CA, USA).
To evaluate the 143B cell adhesion to the decellularized lung scaffold, the decellularized fragments were placed in the 96-well plates and the 143B cells were seeded on top. The cells were allowed to adhere for 10 min at 37 °C and the same protocol as described above was followed to quantify cell adhesion.
Mass spectrometry
Protein extraction
Proteins were reduced and alkylated with 100 mM Tris pH 8.5, 1% sodium deoxycholate, 10 mM tris(2-carboxyethyl)phosphine (TCEP), and 40 mM chloroacetamide for 10 min at 95ºC at 1000 rpm (Thermomixer, Eppendorf). 100 µg of protein were processed for proteomic analysis following the solid-phase-enhanced sample-preparation (SP3) protocol as described by Hughes et al. [37]. Enzymatic digestion was performed with Trypsin/LysC (2 µg) overnight at 37ºC at 1000 rpm. The resulting peptides were cleaned-up and desalted with C18 micro columns and further quantified.
Proteomics data acquisition
Protein identification was performed by nanoLC-MS/MS using an Ultimate 3000 liquid chromatography system coupled to a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany). Peptides were loaded onto a trapping cartridge (Acclaim PepMap C18 100Å, 5 mm x 300 μm i.d., 160,454, Thermo Scientific) in a mobile phase of 2% ACN, 0.1% FA at 10 µL/min. After 3 min loading, the trap column was switched in-line to a 50 cm by 75 μm inner diameter EASY-Spray column (ES803, PepMap RSLC, C18, 2 μm, Thermo Scientific, Bremen, Germany) at 300 nL/min. Separation was achieved by mixing A: 0.1% FA, and B: 80% ACN, with the following gradient: 5 min (2.5% B to 10% B), 120 min (10% B to 30% B), 20 min (30% B to 50% B), 5 min (50% B to 99% B) and 10 min (hold 99% B). Subsequently, the column was equilibrated with 2.5% B for 17 min. Data acquisition was controlled by Xcalibur 4.0 and Tune 2.9 software (Thermo Scientific, Bremen, Germany).
The mass spectrometer was operated in data-dependent (dd) positive acquisition mode alternating between a full scan (m/z 380–1580) and subsequent HCD MS/MS of the 10 most intense peaks from the full scan (normalized collision energy of 27%). ESI spray voltage was 1.9 kV. Global settings: use lock masses best (m/z445.12003), lock mass injection Full MS, chrom. peak width (FWHM) 15s. Full scan settings: 70k resolution (m/z 200), AGC target 3e6, maximum injection time 120 ms. dd settings: minimum AGC target 8e3, intensity threshold 7.3e4, charge exclusion: unassigned, 1, 8, > 8, peptide match preferred, exclude isotopes on, dynamic exclusion 45s. MS2 settings: microscans 1, resolution 35k (m/z 200), AGC target 2e5, maximum injection time 110 ms, isolation window 2.0 m/z, isolation offset 0.0 m/z, spectrum data type profile.
Data analysis
The raw data was processed using the Proteome Discoverer software (Thermo Scientific) and searched against the UniProt database for Homo sapiens Proteome. The Sequest HT search engine was used to identify tryptic peptides. The ion mass tolerance was 10 ppm for precursor ions and 0.02 Da for fragment ions. The maximum allowed for missing cleavage sites was set to 2. Cysteine carbamidomethylation was defined as a constant modification. Methionine oxidation and protein N-terminus acetylation were defined as variable modifications. Peptide confidence was set to high. The processing node Percolator was enabled with the following settings: maximum delta Cn 0.05; decoy database search target FDR 1%, validation based on q-value. Imputation of missing values was performed only when a peptide was detected in at least one of the replicates analyzed. Quantitative evaluation was performed by pairwise comparisons of all detected peptides and the median ratio was used for protein level comparison. Significance assessment was performed using the background-based ANOVA method implemented in Proteome Discoverer 2.2 and multiple comparison adjustment of the p-values was performed.
Protein functional enrichment analysis
The protein functional enrichment analyses for Gene Ontology (GO) terms and Reactome pathways were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.8 bioinformatics tool (https://david.ncifcrf.gov/summary.jsp). The data analysis and visualization of differently expressed proteins were conducted in the R programming language (version 4.1.1) in the RStudio integrated development environment. The GO and pathways plots were achieved using the ggplot2 (version 3.3.5). A basic circle packing chart with parckcircles package (available at https://r-graph-gallery.com/305-basic-circle-packing-with-one-level.html) was used for GO – biological processes. The Venn diagrams were generated with the webtool available at bioinformatics.psb.ugent.be/webtools/Venn/.
Enzyme-linked immunosorbent assay (ELISA)
Levels of EFEMP1 ELISA in mouse serum samples and in the SCR of wild-type, MG-63 cells, 143B cells and EFEMP1-knockdown 143B cells were determined using the Human EFEMP1 ELISA kit (Abcam, ab269552) according to the manufacturer’s instructions.
Kaplan-Meier analysis
The Kaplan-Meier analysis was performed using the R2 database (R2: Genomics Analysis and Visualization Platform – http://r2platform.com) which contains genome-wide gene expression data of OS patient samples (dataset: Mixed OS (Mesenchymal) – Kuijjer – 127 – vst – ilmnhwg6v2). The chondroblastic, fibroblastic and osteoblastic OS patient samples (n = 73) or with metastatic disease (n = 37) in the database were divided into high and low EFEMP1 (ID: 2202) expressions based on scan cut-off.
Statistical analysis
All data are expressed as mean ± standard error of the mean (SEM). Graphics and statistical analysis were performed using GraphPad Prism version 8.0.2 (GraphPad Software, San Diego, CA, USA). Independent variables were analysed by the Mann-Whitney test, whereas Kruskal-Wallis or one-way ANOVA was used for multiple comparisons. Statistical significance was set at the level of p < 0.05. For the Kaplan-Meier analysis of OS patient samples, statistical differences in survival curves were calculated by log-rank (Mantel-Cox) test. Figure illustrations were created with adobe illustrator or BioRender.com.
Results
Osteosarcoma cell-derived secretome induces transcriptome and tissue structural changes similar to a primary growing tumour
As the metastatic microenvironment plays a pivotal role in facilitating the seeding and outgrowth of disseminated tumour cells, we aimed to examine the lung environmental alterations that occur before lung metastasis formation. To establish the role of tumour-derived secreted factors in this process, we used two experimental mouse models. In the first group, animals were implanted subcutaneously with 143B OS cells for primary tumour (PT) formation (hereafter referred to as PT-bearing mice) (Fig. 1A). Tumours were allowed to grow until reaching an average volume of 50–60 mm3, which took approximately 8–10 days (Fig. S1A). To reproduce the continuous secretion of factors by developing primary tumour, a separate group of animals received daily i.p. injections of the secretome (SCR) derived from 143B cells (hereafter referred to as SCR-treated mice) for one week (Fig. 1B). In both conditions, none of the animals showed signs of respiratory distress or weight loss (Fig. S2A-B).
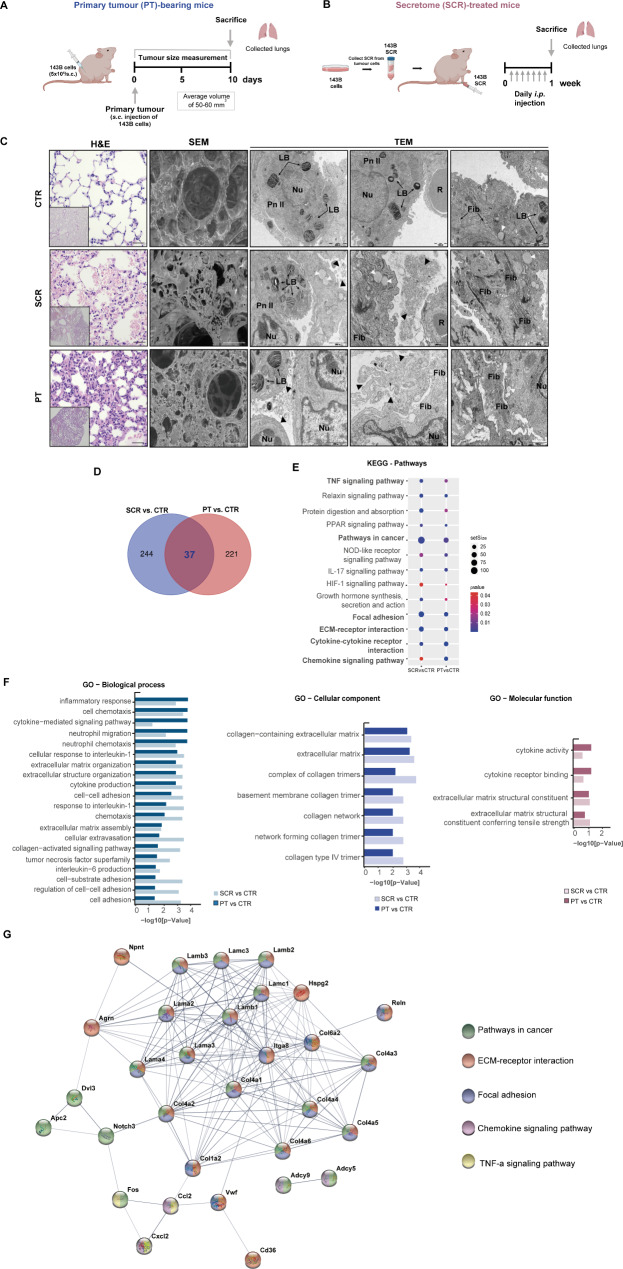
Fig. 1.
Changes in the transcriptome and lung tissue architecture in mice treated with the secretome or with a primary tumour. A Schematic diagram illustrating the s.c. injection of the 143B Luc+ cells into the lower flank of mice. Lung tissue was harvested when the primary tumour (PT) reached a maximum volume of 50–60 mm3 (PT-bearing mice, n = 3–5). B Schematic diagram illustrating the preparation and administration schedule of the 143B cell-derived secretome (SCR) in mice. Animals received daily i.p. injections for 1 week (SCR-treated mice, n = 3–5). Lung tissue was harvested at the end of the treatment. C Representative images of H&E at x200 magnification (Scale bar: 20 μm), SEM (Scale bar: 40 μm), and TEM (Scale bars: 1000 and 2000 nm) of lung tissue sections from untreated mice (CTR), SCR-treated or PT-bearing mice. Black arrowheads: exudate of protein, vesicles, and fragments of the surfactant; White arrowheads: mucous granules (produced by peribronchial glands); LB, lamellar bodies; Nu, nucleus; R, red blood cells; Pn II: pneumocytes type II (alveolar cells); Fib: fibrosis. D Venn diagram of differentially expressed genes (DEGs) in lungs for each pairwise comparison: SCR vs. CTR and PT vs. CTR. E KEGG pathway enrichment analysis of DEGs. Circle sizes denote the number of genes included in a group and the colour indicates the p-value. F Bar plots depicting the manually curated common Gene Ontology (GO) terms found for the two comparison groups. Biological process (BP), cellular component (CC), and molecular function (MF) of altered genes reporting the intersections in lungs from SCR vs. CTR and PT vs. CTR. G Representative protein-protein interaction (PPI) network, constructed with the common DEGs, using the STRING database
The lungs were subsequently harvested for histological and electron microscopic examinations. Histopathological analysis of formalin-fixed paraffin-embedded (FFPE) lung tissue from PT-bearing and SCR-treated mice demonstrated severe damage to the alveolar structure, with significant septum thickening compared to vehicle-treated control (CTR: 7.92 ± 0.40 μm; SCR: 15.79 ± 1.13 μm; PT: 24.25 ± 2.01 μm, p < 0.001), reduced airspace areas, and inflammatory cell infiltration, in comparison to the preserved lung parenchyma from control mice (Fig. 1C). No microscopically detectable lung metastasis was found in the lungs of PT-bearing mice at the endpoint.
Scanning electron microscopy (SEM) images revealed marked changes in the overall alveolar architecture with condensation of the connective tissue framework surrounding the alveolar spaces in the lungs of both PT-bearing and SCR-treated mice. Furthermore, transmission electron microscopy (TEM) images confirmed these observations and showed a disorganized and severely damaged pulmonary structure with leakage of proteins, vesicles, and surfactant (arrowheads) into the alveolar space. Additionally, septum thickening, nucleus with altered morphology, and fibrosis, evidenced by the accumulation of ECM proteins, in comparison with healthy controls.
These findings revealed early onset structural changes in the lung parenchyma, likely induced by the continuous release of distant tumour-secreted factors. Furthermore, the effects resulting from the daily administration of SCR closely resemble those observed in the host with the tumour, indicating a systemic tumour-mediated effect.
To understand the transcriptome alterations underlying lung histological and architectural changes, we performed comparative RNA sequencing-based transcriptome profiling of the lung tissue collected from healthy, SCR-treated, and PT-carrying mice.
Among the 16,138 transcripts, we found a total of 502 differentially expressed genes (DEGs) between the control and experimental groups, using a cut-off of p-value < 0.05 and log fold change (FC)>|1|. Of these, 37 were shared between the two compared groups as shown in the Venn diagram (Fig. 1D). This relatively limited overlap between the two groups may be due to the fact that the SCR was collected from cell monoculture and administered daily in constant volumes. Despite this approach mimicking the systemic release of secreted factors equivalent to a growing primary tumour during the PMN formation, might not fully replicate the secretion dynamics of a growing tumour or the influence of the surrounding stromal microenvironment on its composition.
Even though gene set enrichment analysis (GSEA) identified enriched pathways in cancer, ECM-receptor interaction, focal adhesion, chemokine signalling pathway, and cytokine-cytokine receptor interactions (Fig. 1E). Furthermore, gene ontology (GO) functional enrichment analysis of the common DEGs confirmed their significant contribution to several biological processes (BP) terms, including inflammatory response, cell chemotaxis, cytokine-mediated signalling pathway, neutrophil migration, ECM organization, cytokine production, cell-cell adhesion, cellular extravasation, interleukin-6 production, and cell adhesion (Fig. 1F). The common DEGs were also enriched in cellular components (CC) terms, specifically ECM and collagen, as well as in molecular functions (MF) terms, including ECM structural constituents, cytokine receptor binding and cytokine activity.
Furthermore, a protein-protein interaction (PPI) network analysis of the overlapping 37 DEGs identified 159 edges among 33 nodes (PPI enrichment p-value < 1.0 × 10− 16, using the String platform), with genes such as collagens (Col1, Col4 and Col6), laminins and integrin-alpha-8 as a part of the main cluster of ECM-receptor interactions, focal adhesion and pathways in cancer (Fig. 1G). Collagen, proteoglycans (e.g., versican and hyaluronan) and glycoproteins (e.g., laminins, elastin and fibronectin) are the core matrisome proteins of the ECM that maintain the proper tissue architecture [38]. Changes in ECM remodelling have profound implications for cellular signalling networks, as ECM components can act as pro-inflammatory stimuli and serve as ligands for various molecules and cell surface receptors, such as integrins, likely impacting metastasis formation [38–40]. The transcriptomic data strongly support this hypothesis, as several ECM-related pathways are dysregulated in response to secreted factors from the growing tumour or the administered secretome.
Osteosarcoma-secreted factors cause changes in the lung ECM and the immune and inflammatory landscape during PMN formation
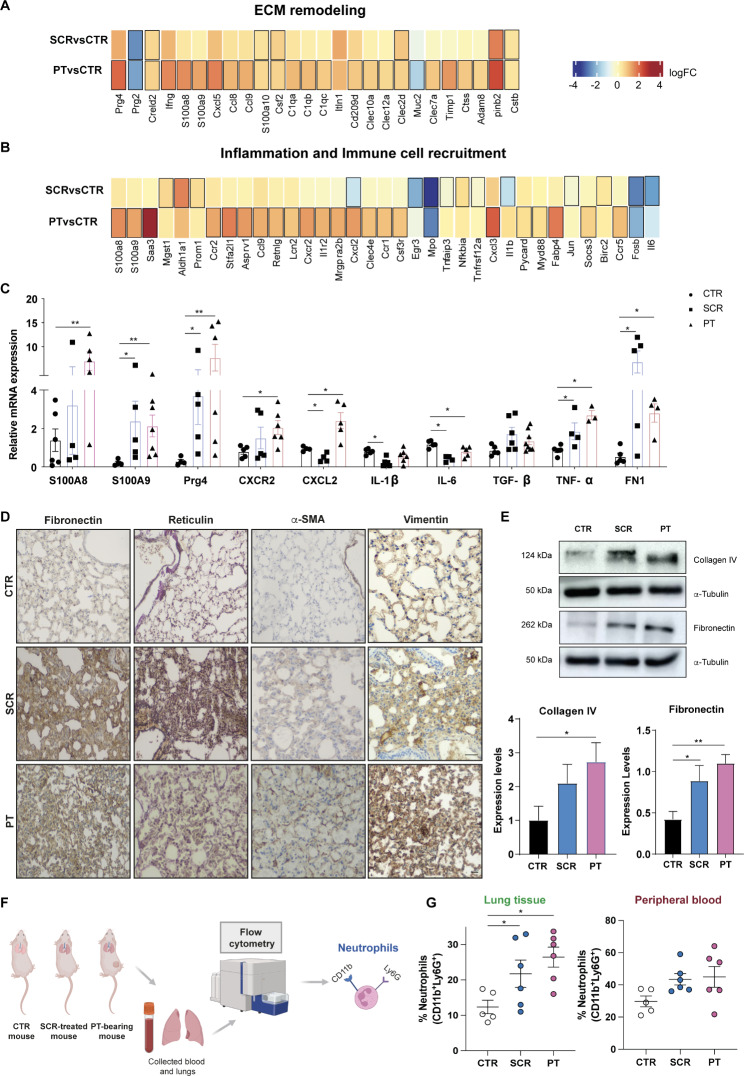
After identifying the core biological pathways through GSEA, we performed a heatmap-based analysis of the relative expression of transcripts linked to ECM remodelling, inflammation, and immune cell recruitment-related pathways between the control and experimental groups. Transcripts with significant expression changes (p-value < 0.05) compared to control mice are marked with a black border. Lung transcriptomic analysis revealed a higher number of significantly dysregulated genes in mice with PT than in those treated with the SCR, although the general trend in gene dysregulation was quite similar (Fig. 2A-B). The heatmap of ECM-related genes revealed 26 deregulated genes (Fig. 2A). Notable findings include the upregulation of proteoglycan 4 (Prg4), known for its anti-inflammatory properties [41, 42], the downregulation of proteoglycan 2 (Prg2), a recognized tumour-suppressor [43], and the upregulation of cysteine-rich with EGF-like domain protein 2 (Creld2), which has implications for tumour progression via fibroblast reprogramming [44]. Other deregulated genes affiliated with the ECM and secreted ECM regulators include C1q complement components of collagen-like structures and C-type lectin-like receptors, the calcium-binding proteins S100A8/A9, the chemokines Cxcl5, Ccl8 and Ccl9, and the colony-stimulating factor (Csf) 2, all of them implicated in promoting cell proliferation, invasion and inflammation [45–49, 51, 52]. Within the heatmap of genes related to inflammation and immune cell recruitment, we found 35 deregulated genes (Fig. 2B), including chemokines, chemokine receptors and also the S100A8/A9, which are involved in the recruitment and activation of myeloid immune cells [50]. Myeloperoxidase (Mpo), an indicator of neutrophil activity [53], was downregulated, as was early growth response 3 (Egr3), a gene with tumour suppressor functions [53, 54]. Upregulated transcripts microsomal glutathione S-transferase (Mgst1) and aldehyde dehydrogenase 1 (Aldh1a1), together with lipocalin-2 (Lcn2), have been implicated in the immunosuppressive function of myeloid-derived suppressor cells (MDSCs) to facilitate tumour progression [55–58], while the fatty acid-binding protein 4 (Fabp4) has been strongly associated with cancer metastasis [49, 59].
Fig. 2.
Lung microenvironmental changes in response to osteosarcoma-secreted factors. A, B Heatmaps of transcripts encoding genes involved in ECM remodelling, inflammation, and immune cell recruitment in lungs from SCR-treated or PT-bearing mice compared with controls. C qRT-PCR analysis of S100A8/A9, Prg4, Cxcr2, Cxcl2, IL-1β, IL6, TGF-β, TNF-α, and FN1 genes in lungs from SCR-treated mice or PT-bearing mice compared with controls. (n = 3–7, per group). D Representative images of fibronectin, reticulin, α-SMA, and vimentin immunostaining at x200 magnification (Scale bar: 30 μm) in lung sections from CTR mice, SCR-treated mice, or carrying a PT. E Western blot analysis of fibronectin and collagen type IV. Expression levels with graphic quantification. F Schematic diagram illustrating the analysis of lung-infiltrating neutrophils and the peripheral blood in mouse models by flow cytometry. G Flow cytometric quantification of infiltrating neutrophils in the lungs and in the peripheral blood of control, SCR-treated and PT-bearing mice (n = 5–6, per group). Data are presented as mean ± SEM from 3–7 independent biological samples. *p < 0.05, **p < 0.01 compared to control lungs (Mann-Whitney test)
Several of the differentially expressed transcripts were confirmed by qRT-PCR in lung tissue lysates, including S100A8/A9, Prg4, Cxcr2, Cxcl2, IL-1β, and IL6, and the results were consistent with RNA-seq data (Fig. 2C). Additionally, we examined the mRNA expression of TGF-β, TNF-α, and fibronectin, known for their immunosuppressive, pro-angiogenic, and ECM regulatory roles, respectively, which are also upregulated in the lungs of PT-bearing and SCR-treated mice compared to the controls.
Immunohistochemical (IHC) analysis of lung tissue sections revealed a marked increase in the deposition of two major ECM components, fibronectin and reticulin fibers, as well as upregulation of the activated fibroblast marker α-SMA and vimentin in both SCR-treated and PT-bearing mice, compared to the control group (Fig. 2D). It was also observed an increase in type IV collagen, (the major component of the basement membrane) by western blot (WB), as well as of fibronectin (Fig. 2E). These findings support the transcriptomic evidence of lung ECM remodelling during PMN formation.
After identifying DEGs enriched in biological processes related to the recruitment of myeloid cells, we conducted a flow cytometry analysis of infiltrating neutrophils and other immune cells (macrophages, eosinophils, monocytes and B-lymphocytes) in the lungs and peripheral blood of control, SCR-treated, and PT-bearing mice (Fig. 2F). The results showed a significant increase in the percentage of infiltrating neutrophils (CD11b+ Ly6G+) in the lungs of PT-bearing or SCR-treated mice compared to age-matched untreated mice (Fig. 2G). A similar trend was observed in the peripheral blood, albeit without reaching statistical differences. Regarding other immune cell populations (monocytes, macrophages, eosinophils and B-lymphocytes), no significant differences were observed in either the lungs or peripheral blood among the different experimental groups (Fig. S3A-B).
Depending on the microenvironment, neutrophils can acquire an anti-tumorigenic N1 or a pro-tumorigenic N2 phenotype [60]. In our study, we could not identify the phenotype of lung-infiltrating neutrophils, but the increased mRNA levels of TGF-β, a major regulator of neutrophil polarization, suggest an N2-like phenotype. Moreover, the transcriptomic analysis revealed an upregulation of Lcn2, a secreted glycoprotein produced by N2-neutrophils and implicated in promoting metastasis [61, 62], simultaneously with a marked downregulation of Mpo in the premetastatic lungs, providing additional support for this hypothesis.
Altogether, these findings suggest that tumour-secreted factors instigate inflammatory signalling pathways in the lung microenvironment with concurrent ECM remodelling and neutrophil infiltration as part of PMN formation.
Activated lung fibroblasts in premetastatic lungs drive fibronectin and collagen deposition and favour the adhesion of OS cells to the lung ECM
Since fibroblasts are the predominant cells in the lung interstitium and the major producers of ECM components, we hypothesized that tumour-secreted factors trigger the activation of stromal fibroblasts for ECM remodelling.
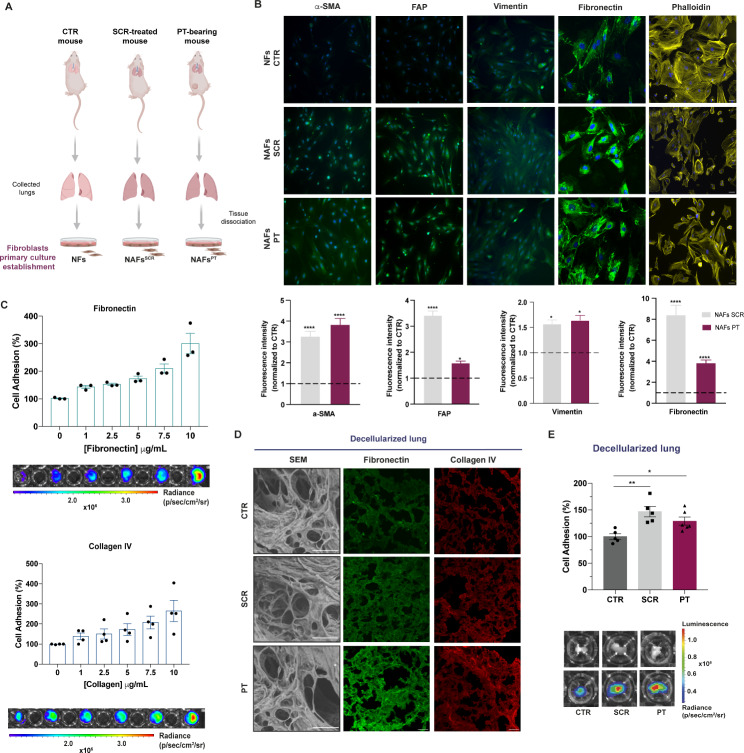
To address this, we performed a phenotypic profiling analysis of lung fibroblasts directly isolated from freshly excised lungs of SCR-treated and PT-bearing mice, as schemed in Fig. 3A. Fibroblasts from age-matched healthy mice were used as controls and were termed as normal fibroblasts (NFs). Immunostaining showed an upregulation of α-SMA and FAP, two classical markers of activated fibroblasts, as well as of the intermediate filament vimentin, in fibroblasts from SCR-treated animals or PT-bearing animals, hereafter referred to as normal activated fibroblasts, NAFsSCR or NAFsPT, respectively. The strong immunoreactivity for fibronectin in NAFsSCR and NAFsPT, further supports the contribution of stromal fibroblasts to lung ECM remodelling, as previously described. Furthermore, staining with phalloidin, a marker for F-actin filaments, revealed alterations in actin organization, characterized by an irregular and disorganized shape and shortening of filament structures (Fig. 3B) in fibroblasts from PT-bearing and SCR-treated animals. These phenotypic changes were accompanied by an increase in cell motility, as assessed by a wound healing assay (Fig. S4A-B).
Fig. 3.
Increased deposition of fibronectin and collagen by activated lung fibroblasts favour the adhesion of OS cells to the lung ECM. A Schematic diagram of the establishment of the primary cultures of fibroblasts isolated from the lungs of untreated (NFs), SCR-treated (NAFsSCR) and PT-bearing mice (NAFsPT). B Representative image of immunofluorescence staining for α-SMA, FAP, vimentin, fibronectin and phalloidin at x20 magnification (Scale bar: 50 μm) in primary cultures of fibroblasts from CTR, SCR-treated or PT-bearing mice. Immunofluorescence quantification of α-SMA, FAP, vimentin and fibronectin (n = 3, per group). C Relative adhesion of 143B cells to increasing concentrations of fibronectin and collagen type IV ranging from 1 to 10 µg/mL (n = 3–4, performed in triplicate). Representative bioluminescence images of cell adhesion (143B cells) to different concentrations of fibronectin and collagen IV. D Representative SEM images (Scale bar: 50 μm) of decellularized lung sections and fibronectin and collagen immunostaining at x20 magnification (Scale bar: 50 μm) in decellularized lung sections from CTR mice, SCR-treated mice, or carrying a PT. E Relative adhesion of 143B cells to decellularized lung sections from CTR, SCR-treated, or PT-bearing mice (n = 5). Representative pictures of the decellularized fragments in the wells (upper row) and bioluminescent images of adhered cells (lower row). The bioluminescent signal is represented as radiance (p/s/cm2/sr). Data are presented as mean ± SEM. *p < 0.05 and ****p < 0.0001 are significantly different when compared to NFs (Kruskal-Wallis (B)); *p < 0.05 and **p < 0.01 when compared to healthy decellularized lungs (One-way ANOVA (E))
Given that fibronectin is a cell-adhesive glycoprotein known to promote tumour cell adhesion, we hypothesized that secretome-induced upregulation of fibronectin might promote further lung colonization by OS cells. To address this, we performed a cell adhesion assay with 143B cells on fibronectin-coated plates using coating concentrations ranging from 1 to 10 µg/mL. The results showed a gradual increase in the percentage of adherent cells with increasing fibronectin concentrations. Similar results were observed on collagen IV-coated plates, underscoring the role of these proteins as substrates for cell attachment (Fig. 3C).
Encouraged by these findings, we evaluated the adhesion of 143B cells to decellularized lung fragments from SCR-treated or PT-bearing mice. Immunofluorescence staining confirmed the increased deposition of fibronectin and collagen IV in decellularized fragments of animals treated with the SCR or with a PT (Fig. 3D), in agreement with IHC and WB data (Fig. 2D, E). We then seeded 143B cells on plates coated with decellularized lung scaffolds and allowed them to adhere for 10 min. Our results showed a significant increase in the percentage of 143B cells attached to the lung scaffolds of both SCR-treated (p < 0.01) and PT-bearing mice (p < 0.05), representing an approximately 50% increase compared to control animals (Fig. 3E). Furthermore, immunostaining of decellularized fibroblast cell sheets revealed an increase in staining intensity for fibronectin and collagen IV (Fig. S5), confirming the contribution of activated fibroblasts to ECM remodelling and cell adhesion.
Tumour secretome-induced microenvironmental changes in pre-metastatic lungs promote and accelerate lung metastasis formation
Up to this point, our data showed the impact of the OS cell secretome on lung PMN formation. To confirm whether this effect promotes and/or accelerates lung metastasis formation, we used an experimental metastasis model in mice. Animals were treated with the 143B-derived secretome daily for one week, followed by the i.v. injection of tumour cells into the tail vein (Fig. 4A). Non-preconditioned mice were used as the controls (Fig. 4B). Lung metastasis formation was monitored weekly for over 60 days using bioluminescence imaging (BLI).
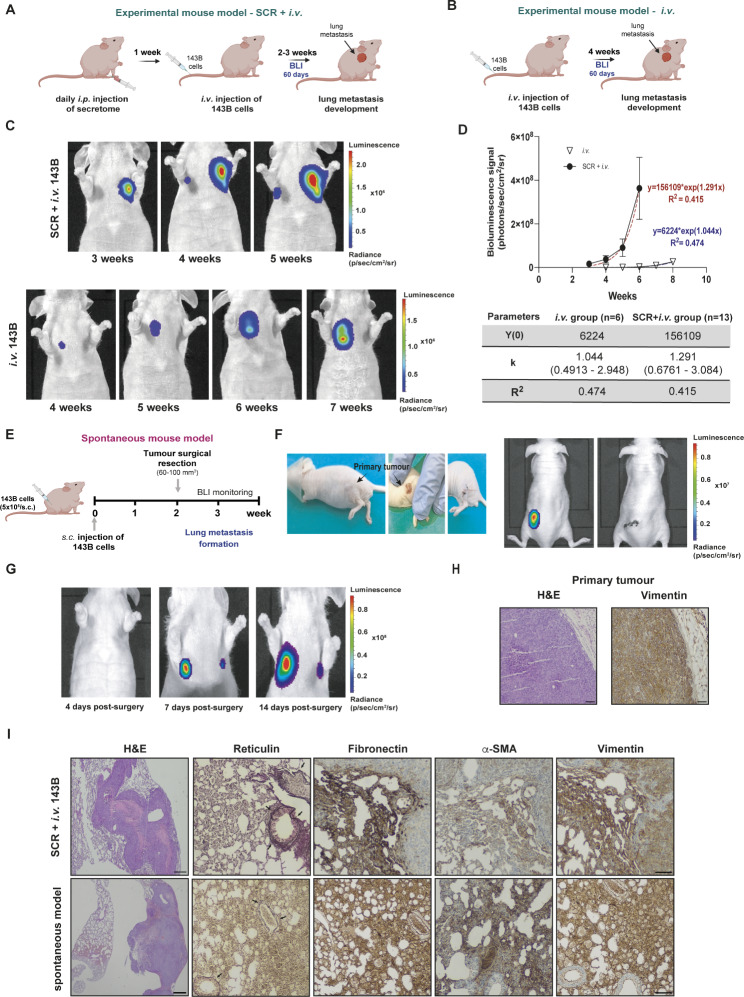
Fig. 4.
Osteosarcoma-induced PMN formation promotes and accelerates the formation of lung metastasis. A Schematic diagram of the experimental model of lung metastasis. Mice were treated with 143B cells-derived secretome (SCR) for 1 week, followed by i.v. administration of 143B Luc+ cells into the tail vein (SCR + i.v. group). B Schematic diagram of the experimental model of lung metastasis. Mice received only the i.v. injection of 143B Luc + cells without pre-treatment with the SCR (i.v. group). C Representative bioluminescence images of lung metastasis formation in pre-treated (SCR + i.v. group) and untreated (i.v. group) mice with secretome before the i.v. injection of 143B cells. D Exponential fitting of the bioluminescence signals (photons/second) of metastatic lesions over time in the i.v. group (∇ n = 6) and the SCR + i.v. group (• n = 13 mice), and corresponding kinetic parameters. E Schematic diagram of the spontaneous metastatic mouse model. Animals were injected subcutaneously with the 143B cells. After reaching a volume of 60–100 mm3, the tumour was excised, and the animals were monitored by BLI for lung metastasis formation. F Images of the surgical resection of a primary tumour with a volume of 60 mm3 and bioluminescence images before and after the excision of the tumour. G Representative bioluminescence images at 4, 7 and 14 days after surgical resection of the primary tumour. H Histological H&E images at x100 magnification (Scale bar: 30 μm) and immunostaining for vimentin at x100 magnification (Scale bar: 30 μm) of the resected tumour. I Histological H&E images at x40 magnification (Scale bar: 20 μm) and IHC staining for reticulin (arrowed), fibronectin, α-SMA, and vimentin at x100 magnification (Scale bar: 30 μm) of lung metastatic lesions in both experimental and spontaneous mouse models. Data are presented as mean ± SEM
All preconditioned animals developed lung metastasis, detected in the second to third weeks after cell inoculation, which rapidly progressed to larger and multilobular lesions. In contrast, only 55% of the untreated group, developed metastasis by the fourth week, mostly unilobular, with slower growth kinetics, never reaching the size of the treated group (Fig. 4C). Parameters estimated from the exponential fitting of the bioluminescent signals confirmed a faster growth rate of metastatic lesions in animals pre-treated with the OS cells secretome (Fig. 4D). The development of metastasis in some untreated animals was probably due to the highly aggressive nature of the143B cells. However, the metastatic rate was substantially lower and the lesions were smaller and less invasive than those in the preconditioned group.
We also established a spontaneous metastatic animal model to assess whether PMN formation induced by an engrafted primary tumour dictate the innate propensity of OS to metastasize to the lung. A primary tumour was induced by subcutaneous injection of 143B cells into the lower flank and allowed to grow until it reached a maximum volume of 60–100 mm3 (Fig. 4E). The tumour was then surgically excised and the animals were monitored for lung metastasis formation. Post-operative BLI images confirmed the complete removal of the primary tumour (Fig. 4F) and the absence of lung metastasis or metastasis to other organs. Four days post-surgery, there were no signs of lung metastasis; however, a week later, two nodules were detected, which progressively evolved over the following two weeks (Fig. 4G). These results suggest that, at the time of surgery, tumour cells were already disseminated in the circulation at the time of surgery and were able to colonize the permissive lung microenvironment induced by the primary tumour, allowing for their survival and metastatic outgrowth. The spontaneous formation of distant lung metastasis further confirms the organotropism of OS in this organ, which is thought to be achieved by factors secreted by the engrafted OS cells.
Histopathological examination confirmed the high-grade malignancy in both primary OS (Fig. 4H), and lung metastasis in both the experimental and spontaneous models (Fig. 4I), as evidenced by marked nuclear pleomorphism, the high mitotic rate, and intralesional necrotic areas. The Gordon-Sweet staining of lung sections, using silver impregnation, revealed an increased deposition and rearrangement of fibrillar collagens surrounding the metastatic lesions and around the bronchi and blood vessels, consistent with a fibrotic response.
Additionally, IHC showed strong immunoreactivity for fibronectin and α-SMA in the stromal regions of the periphery of the neoplastic lesions, which revealed the presence of activated fibroblasts in the lung. The mesenchymal marker vimentin, also known as fibroblast-intermediate filament, exhibited strong immunoreactivity in stromal and metastatic cells, confirming the mesenchymal origin of the latter.
Overall, these findings confirmed that lung alterations induced by the secretome or grafted OS cells create a favourable environment for the survival and outgrowth of metastatic cells and undergo continuous remodelling.
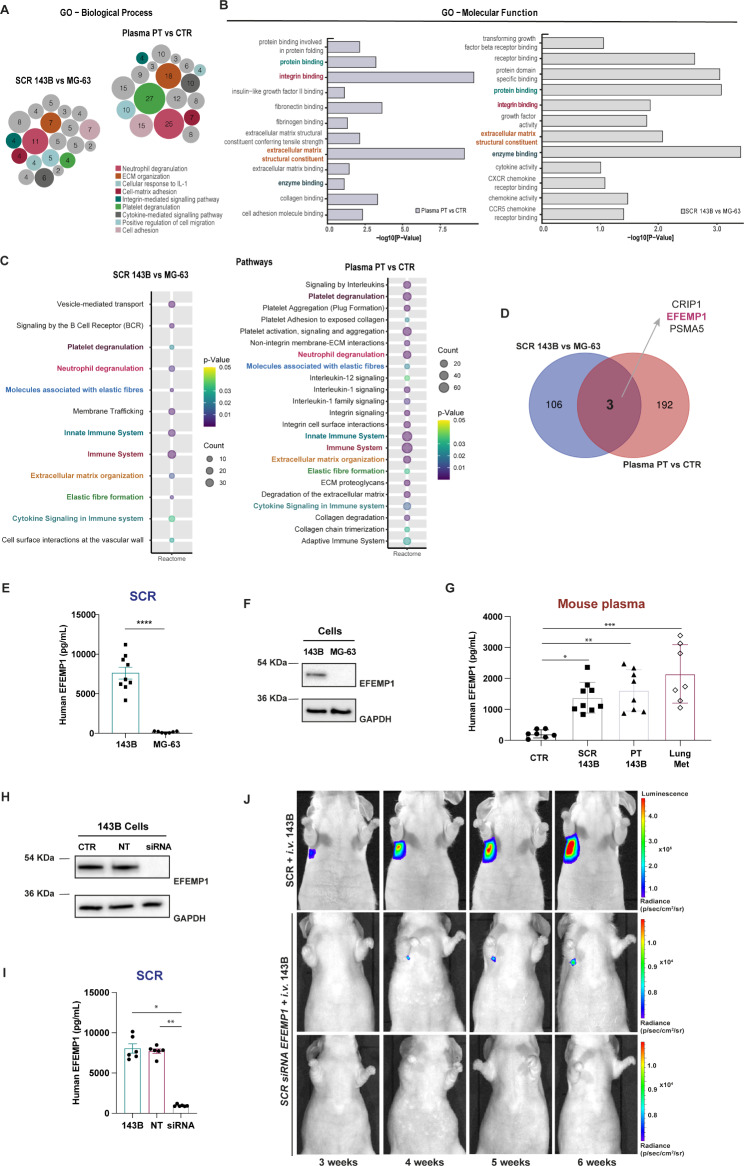
Analysis of the proteomic profile of cell secretome and mouse plasma identified EFEMP1 as a potential prognostic biomarker in osteosarcoma
Since the effects of daily administration of SCR on a premetastatic lung were quite similar to those elicited by a local primary tumour, we performed a comparative mass spectrometry-based label-free quantitative proteomic analysis of the 143B SCR and the plasma from PT-bearing mice to identify potential common mediators predictive of lung metastasis formation. Secretome collected from a non-metastatic OS cell line (MG-63) [63] and plasma from healthy mice were used as controls. We confirmed the non-metastatic potential of MG-63 cells in mice, as none of the animals i.v. injected with these cells developed metastases (Fig. S6A).
A list of differentially expressed proteins (DEPs) of 143B versus MG-63 OS cells and plasma from healthy mice versus PT-bearing mice was generated using an FDR-adjusted p-value ≤ 0.05. A total of 2,595 proteins were identified in the secretome of OS cells, with 109 proteins differentially expressed between the 143B and MG-63. In the mouse plasma, we considered only proteins of human origin to focus on what was secreted by human OS-induced tumours. Out of these, we identified 195 differentially expressed between the plasma samples of PT-bearing mice and healthy controls.
GO enrichment analysis was performed on DEPs between the two pairwise comparisons to identify shared biological processes and molecular functions. The largest number of identified proteins was particularly enriched in neutrophil degranulation, platelet degranulation, ECM organization, and cell adhesion (Fig. 5A-B). Reactome enrichment analysis identified 8 common overlapping pathways engaged in platelet degranulation, neutrophil degranulation, molecules associated with elastic fibres, innate immune system, immune system, ECM organization, elastic fibre formation and cytokine signalling in the immune system (Fig. 5C). All of these pathways are known to be involved in PMN formation [64–67]. The Venn diagram in Fig. 5D identified three overlapping DEPs between the two pairwise comparisons, which include the epidermal growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1, also called fibulin-3), cysteine-rich protein 1 (CRIP1) and proteasome subunit alpha type-5 (PSMA 5). Among these proteins, EFEMP1 was the most abundant in both the SCR and mouse plasma, exhibiting consistently high-intensity peak values and greater protein coverage across all replicates. EFEMP1 is an ECM glycoprotein broadly expressed in various tissues during development and adulthood. As a crucial component of the basement membrane, EFEMP1 plays a crucial role in preserving the structural integrity and stability of the ECM [68]. Considering the ECM alterations in the lungs of mice challenged with the SCR or carrying a PT, we reasoned that secreted EFEMP1 could play an important role in this process. To address this, we validated our proteomic approach by measuring protein expression and secreted levels of EFEMP1 by OS cells in vitro.
Fig. 5.
Proteomic analysis identified EFEMP1 as a potential metastatic-related biomarker in osteosarcoma. A, B Gene ontology analysis (GO) output. Biological process (BP) and molecular function (MF) of differently expressed proteins (DEPs) in the two pairwise-comparison groups (SCR 143B vs. MG-63; Plasma PT vs. CTR). Circle sizes in BP denote the number of genes involved in the process. C Reactome pathway enrichment analysis of identified proteins in the two pairwise- comparison groups. Circle sizes denote the number of genes included in a group and colour indicates the p-value. The common pathways are highlighted. D Venn diagram showing specific and common proteins among the two pairwise groups: SCR 143B vs. MG-63 and Plasma PT vs. CTR. E EFEMP1 levels in the SCR of the metastatic 143B and non-metastatic MG-63 OS cells. F Representative western blot of EFEMP1 in the metastatic 143B and non-metastatic MG-63 OS cells. G Plasma levels of EFEMP1 in control mice (CTR), mice treated with the 143B SCR, bearing a primary tumour (PT) or with lung metastasis (Lung Met). H Representative western blot of EFEMP1 expression in 143B cells (CTR), non-targeting (NT) and siRNA knockdown of EFEMP1 cells. I EFEMP1 levels in the SCR of 143B cells, NT and siRNA EFEMP1-knockdown cells. J Representative bioluminescence images of mice treated with the SCR of control 143B (SCR + i.v. group) or EFEMP1-knockdown cells (SCR siRNA EFEMP1 + i.v. group) followed i.v. injection of 143B-Luc+ cells. Data are presented as mean ± SEM, from 7 to 8 independent experiments. **** p < 0.0001 were significantly different when compared with the SCR from MG-63 (unpaired t-test (E)); *p < 0.05, **p < 0.01 and ***p < 0.001 were significantly different when compared with the values present in the plasma from healthy mice (Kruskal-Wallis test (G); *p < 0.05, **p < 0.01 were significantly different when compared with 143B SCR (Kruskal-Wallis test (I))
ELISA and western blot analysis confirmed the exclusive expression and secretion of EFEMP1 by 143B cells, with negligible levels observed in the non-metastatic MG-63 cell line, thereby supporting the proteomic data (Fig. 5E, F). Moreover, EFEMP1 was detected in the plasma of animals treated with the secretome or carrying a PT, as well as in those with established lung metastasis, with higher levels observed in the latter (Fig. 5G). The use of a human EFEMP1 ELISA kit points to 143B cells as a source of the plasmatic EFEMP1 levels. Importantly, this protein is involved in 3 out of the 10 common biological processes, specifically ECM organization, molecules associated with elastic fibres and elastic fibre formation, suggesting that it may contribute to ECM remodelling during metastasis.
To evaluate the functional effects of secreted EFEMP1 in lung metastasis formation, we knocked down 143 cells for EFEMP1 and collected the SCR. The siRNA knockdown resulted in effective protein silencing and a robust 90% reduction in EFEMP1 secretion, as confirmed by Western blot and ELISA, respectively (Fig. 5H, I). Non-targeting siRNA did not affect EFEMP1 expression or secretion. A new group of animals was then preconditioned with the SCR of EFEMP1-silenced cells and then injected into the tail vein with 143B-Luc + cells, as previously described. Remarkably, this approach resulted to the complete prevention of lung metastasis formation in 80% (4/5) of the animals throughout the 40-day monitoring period by BLI. The single case of metastasis that did occur was observed at later time-points and exhibited a slower progression rate compared those received the whole-secretome (Fig. 5J).These data demonstrate that EFEMP1 is indeed required to foster a permissive PMN to support the colonization, survival, and proliferation of incoming cells.
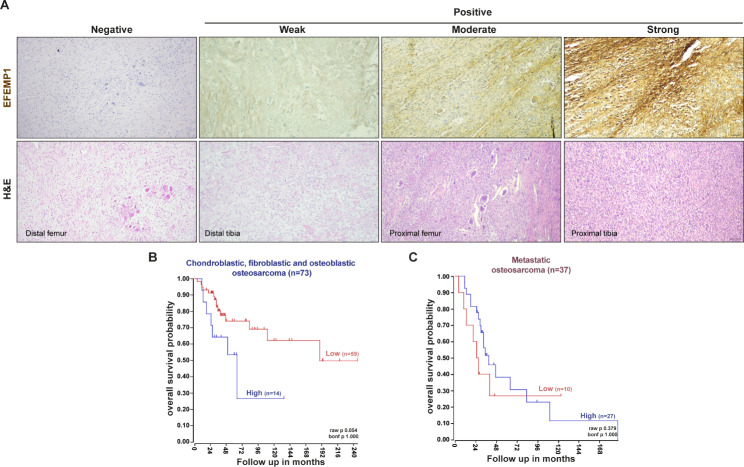
To explore the potential prognostic significance of EFEMP1 expression in OS patients, we conducted a retrospective study on a small cohort of 29 patients without evidence of metastasis at the time of diagnosis. Archived paraffin-embedded biopsy specimens of the primary tumour were immunohistochemically stained for EFEMP1 to investigate its association with the occurrence of lung metastasis during the follow-up period. Of the 29 patients, 12 (41.4%) experienced lung metastasis, and 50% of these patients had positive biopsy specimens for EFEMP1 with a disease-free interval (DFI) of 38.5 months (range: 6–77 months). The other 6 patients who tested negative for EFEMP1 had a longer DFI of 44 months (range: 15–159 months). Among the 17 patients who did not develop lung metastasis, only 6 (35%) showed positive staining for EFEMP1. Importantly, patients with EFEMP1-positive biopsies had a higher mortality rate of 58% compared to those with negative biopsies, whose mortality rate was 29%. Representative images of H&E and immunostaining for EFEMP1 in biopsy samples with negative, weak, moderate and strong intensities are shown in Fig. 6A. Positive staining was primarily observed in the cytoplasm and membrane of OS cells.
Fig. 6.
EFEMP1 expression in primary tumours correlates with poor prognosis in OS patients. A Representative images of EFEMP1 staining and H&E in biopsy samples of high-grade OS patients at x100 magnification (Scale bar: 50 μm). B, C Kaplan-Meier analysis of overall survival in chondroblastic, fibroblastic and osteoblastic OS patient samples (n = 73 patients) and with metastatic disease (n = 37 patients). Scan cut-off was used to group samples into high (blue) and low (red) EFEMP1 expressions. p-values were determined by a log-rank test
Due to the small sample size in our study, we were unable to perform a reliable Kaplan-Meier survival analysis. Therefore, we used the recognized R2 bioinformatic platform to conduct a univariate analysis, which revealed a significant correlation between high EFEMP1 expression and poorer overall survival (246 vs. 135 months) in high-grade conventional OS patients (raw p-value = 0.05, Fig. 6B). However, in the case of patients with lung metastases, we did not find a significant correlation between EFEMP1 expression and the overall survival (raw p-value = 0.379, Fig. 6C). These findings suggest that EFEMP1 upregulation in primary tumours is associated with poor prognosis in OS patients, particularly those with high-grade conventional OS. However, further studies using larger sample sizes are required to confirm these results and understand the underlying mechanisms of EFEMP1 in cancer progression and patient outcomes.
Discussion
In this study, we aimed to understand the mechanisms by which OS cells reprogram the lung microenvironment to support colonization and outgrowth of disseminated tumour cells. Employing two experimental mouse models and a multi-omics approach, we demonstrated that OS cell-derived secreted factors instigate a permissive lung PMN for incoming tumour cells, comprising ECM remodelling, fibroblasts activation, neutrophil recruitment and shaping of the inflammatory environment. These alterations, observed in animals carrying a primary tumour or treated with SCR, highlight the pivotal role of OS-secreted factors in setting up the lung metastatic process.
The transcriptome data identified several DEGs enriched in related biological processes, indicating ECM remodelling, inflammatory response, cell adhesion and chemotactic signals as essential steps. IHC analysis confirmed the increased deposition of fibronectin and fibrillar collagen in the lungs of both experimental mouse models, a result that was further verified in decellularized lung fragments. These proteins, in addition to forming the ECM backbone, act as anchor points for cell adhesion molecules, like β1 and α6 integrins, as well as CD44, all of which are expressed in 143B OS cells (Fig. S7). Indeed, the increased attachment of 143B cells to decellularized lung scaffolds, confirms the pivotal role of fibronectin and collagen in mediating cell adhesion, regarded as a critical step for organ colonization (Fig. 3C, E). The remodelling of the ECM is also implicated in metastatic organotropism through organ-specific integrin-ECM interactions. Exosomal α6β4 and α6β1 integrins have been shown to dictate the exosome adhesion to ECM components and lung resident fibroblasts, resulting in stromal remodelling to host metastatic cells [69]. Although, we have not explored the individual contribution of exosomes to PMN formation, recent studies from our group confirmed that they express β1 and α6 integrins as their parental cells and have an intrinsic homing ability for homotypic lung metastasis [27].
Fibroblasts are the main producers of ECM components, both in homeostasis and in response to injury. In tumours, they are the major players in dysregulated collagen turnover and fibronectin deposition, manifested as desmoplasia [70–72]. Our findings confirmed a fibroblast-to-myofibroblast transition during the PMN, with concomitant deposition of fibronectin and collagen, that persists throughout metastatic progression. It is worth noting that these fibroblasts were isolated directly from tumour-free lung tissue without any intervening culture steps that might have impacted gene expression, further confirming the ability of fibroblasts to functionally adapt to the evolving environment. Cancer-associated fibroblasts (CAFs) are the most prominent stromal cells and serve as positive regulators of tumour growth at the primary tumour site by reprogramming the local microenvironment [73–75]. Similarly, metastasis-associated fibroblasts (MAFs) have been found to promote metastatic outgrowth by engaging in inflammatory-mediated communication with cancer cells [76, 77]. These findings were obtained in models with established lung macrometastases, whereas in our study, the fibroblasts phenotypic changes were observed in an incredibly early stage before tumour cell colonization.
The cytokine TGF-β is a major inducer of myofibroblast differentiation through autocrine and paracrine mechanisms, resulting in the secretion of additional growth factors, cytokines and chemokines involved in a complex integration of signals promoting the recruitment of BMDCs, angiogenesis, immunosuppression, and ECM remodelling [78, 79]. Transcriptome and RT-PCR analyses confirmed the expression of TGF-β and other fibroblast activation inducers (e.g. TNF-α and CCl2) in premetastatic lungs, suggesting the fibroblast contribution in sustaining an inflammatory response. These findings are consistent with a study by Mazumdar et al. [80] on highly metastatic OS cell lines, including 143B, where they found that OS-derived EVs induced lung myofibroblast differentiation through the activation of the TGF-β1 and SMAD2 pathways. Flow cytometric analysis showed a significant increase in the infiltration of CD11b+Ly6G+ neutrophil (but not of other immune cells) in the pre-metastatic lungs of both models compared with age-matched controls. Similar findings were reported by Hiratsuka et al., who showed that TNF-α-induced S100A8 and A9 signalling promotes the expression of serum amyloid A3 (SAA3) and the recruitment of CD11b+ myeloid cells to premetastatic lungs [50, 81]. A large number of clinical studies have correlated high levels of intratumoral and systemic neutrophils with poorer disease prognosis, attributed to the release of pro-angiogenic and tumour-growth factors and the suppression of immune responses [82–84]. A few studies have reported the accumulation of neutrophils in future metastatic sites, but their role at this early stage remains elusive as both pro- and anti-metastatic functions have been described [26, 85–87]. As part of the innate immune system, neutrophils are recruited to sites of inflammation driven by cytokines and chemotactic factors to modulate the inflammatory process. Transcriptomic and qPCR analyses identified expression of several members of the CXC and CC chemokines and S100A8/A9 that are chemotactic for neutrophils, which could explain their accumulation in premetastatic lungs, but not their functional significance. Nevertheless, the upregulation of TGF-β and the marked downregulation of neutrophil MPO suggest a pro-tumorigenic role rather than anti-metastatic protection. A limitation of this study is the use of immunocompromised mice, which does not allow us to assess the contribution of neutrophils to the suppression of T cell-mediated responses. However, inhibition of Ly6G + cell mobilization has been shown to prevent lung metastasis formation in both immunocompetent and immunodeficient mice, suggesting a T cell-independent effect, at least in the early stages [86]. However, further studies are needed to clarify the pro-metastatic contribution of neutrophils to PMN formation.
The follow-up animal studies confirmed that systemic remodelling of the lung microenvironment in the SCR-treated and PT-bearing mice promoted and accelerated lung metastasis formation, suggesting that a pre-existing growth-permissive niche is crucial for the incoming of metastatic cells.
Comparative analysis of differentially secreted proteins in the SCR of 143B cells and plasma samples of PT-bearing mice, identified common significantly enriched GO terms related to ECM organization, cell-matrix adhesion, neutrophil and platelet degranulation, and cytokine-mediated signalling. These findings are consistent with lung transcriptomic GO terms and, more importantly, with the lung microenvironmental changes previously described.
The glycoprotein EFEMP1 was one of the enriched proteins that was found in the 143B cell SCR, and in the plasma of mice with primary OS, lung metastasis or treated with the 143B cell-derived SCR in a tumour-free setting, suggesting a driving oncogenic role for lung metastasis in a premetastatic stage. The suppression of lung metastasis formation observed in 80% (4/5) of mice preconditioned with EFEMP1-depleted SCR, strongly supports this hypothesis, and importantly confirm the crucial role of this secreted protein in providing a permissive microenvironment, which is required for the subsequent survival and outgrowth of OS cells in the lung.
As already mentioned, EFEMP1 plays a central role in maintaining the structural integrity of the ECM, organizing the ECM scaffold and in promoting cell-ECM adhesion in normal connective tissue and disease [88]. This protein is upregulated in some tumours and has been described as a novel soluble signal strongly associated with the malignant progression in gliomas, pancreatic, ovarian, and bladder cancer, as well as OS, by promoting cell proliferation, MMP-induced invasion, migration, EMT, and angiogenesis [89–93]. Although not explored, we reasoned that EFEMP1 is involved in ECM remodelling and/or contributes to cell-matrix adhesion, which are integral parts of the premetastatic process, as previously observed in our study. Moreover, EFEMP1 is part of the core effectors of these biological processes. These findings point to EFEMP1 as a putative therapeutic target and a novel plasma biomarker with added value in predicting the risk of lung metastasis in OS. However, further mechanistic studies are warranted to understand the significance of EFEMP1-dependent SCR in favouring the metastatic potential of OS cells and to dissect the interplay between tumour and stromal cells mediated by EFEMP1.
In clinical studies, the small size of our patient cohort precluded us from establishing a definite causal relationship between EFEMP1 expression in primary tumour and the development of lung metastases. Nevertheless, we observed that patients with EFEMP1-expressing tumours have poorer overall survival, suggesting an association with tumour aggressiveness, which is substantiated by the Kaplan-Meier survival analysis. Further research with larger patient cohorts is needed to confirm this hypothesis and gain a more comprehensive understanding of EFEMP1 involvement in the disease.
Conclusions
In summary, our study sheds light on systemic-induced lung microenvironmental changes that precede metastatic spread in OS. Integration of our data uncovers neutrophil infiltration and the functional contribution of stromal-activated fibroblasts in ECM remodelling for tumour cell attachment as early pro-metastatic events, which may hold therapeutic potential in preventing or slowing metastatic spread. Moreover, we identified EFEMP1, a secreted protein by OS cells, as a potential driver of lung metastasis. It not only holds promise as a therapeutic target but also as a plasma biomarker with added value in predicting the risk of lung metastasis in OS.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Teresa Ribeiro-Rodrigues from the Microscopy and Bio-Imaging Lab (iLAB) and Isabel Silva & Sandra Silva from the Flow Cytometry Functional Unit Core Facility.
Abbreviations
- BLI
Bioluminescence imaging
- BMDCs
Bone marrow-derived cells
- BP
Biological Process
- CAFs
Cancer-associated fibroblasts
- CC
Cellular components
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DEGs
Differently expressed genes
- DEPs
Differentially expressed proteins
- DFI
Disease-free interval
- ECM
Extracellular matrix
- ELISA
Enzyme-linked immunosorbent assay
- EVs
Extracellular vesicles
- FDR
False discovery rate
- FFPE
Formalin-fixed paraffin-embedded
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- i.p.
Intraperitoneal
- i.v.
Intravenous
- IHC
Immunohistochemical
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAFs
Metastasis-associated fibroblasts
- MF
Molecular Function
- NAFs
Normal activated fibroblasts
- NFs
Normal fibroblasts
- OS
Osteosarcoma
- PMN
Premetastatic niche
- PPI
Protein-protein interaction
- PT
Primary tumour
- RT
Room Temperature
- SCR
Secretome
- SEM
Scanning electron microscopy
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- TEM
Transmission electron microscopy
- WB
Western blot
Authors’ contributions
S.A., L.S., G.S.-R. and H.F. performed experiments, acquisition, analysis and interpretation of data. N.L., R.C., M.Z., A.P., P.T., R.A. and T.M.-M. performed experiments. M.A. performed bioinformatics analysis. A.S.R. performed proteomic analysis. H.G., J.M.C. and A.J.A. assisted with data analysis. S.A. wrote the manuscript. C.M.G. designed and supervised the study, revised the manuscript and obtained financial support. All authors reviewed the manuscript and approved its content.
Funding
This work was supported by the Portuguese Foundation for Science and Technology (FCT) through the project PTDC/BTM-SAL/4451/2020 and STRATEGIC PROJECTS (UIDB/04539/2020 and UIDP/04539/2020). S.A. is a PhD fellow of the FCT (PD/BDE/142929/2018). L.S. is a PhD fellow of the FCT (PD/BDE/150707/2020). G.S.-R. is a PhD fellow of the FCT (UI/BD/154407/2022).
Data Availability
The datasets generated in this study are available in the following databases:
RNA-Seq data: GEO database [GSE216744] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216744).
Protein identification: PRIDE database [PXD040814].
Declarations
Ethics approval and consent to participate
All animal experiments were approved by the Animal Welfare Committee of the Faculty of Medicine of the University of Coimbra and conducted following the European Community directive guidelines for the use of animals in the laboratory (2010/63/EU) transposed to the Portuguese law in 2013 (Decreto-Lei 113/2013). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Ethics Committee of the Centro Hospitalar e Universitário de Coimbra (CHUC) (CHUC-021/19). Written informed consent was provided by all patients or their legal guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to Success. J Clin Oncol. 2015;33(27):3029–35. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, Fiedorowicz M, et al. Molecular biology of osteosarcoma. Cancers. 2020;12(8):2130–57. doi: 10.3390/cancers12082130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheumatol Therapy. 2017;4(1):25–43. doi: 10.1007/s40744-016-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moukengue B, Lallier M, Marchandet L, Baud’huin M, Verrecchia F, Ory B, et al. Origin and therapies of Osteosarcoma. Cancers (Basel) 2022;14(14):3503. doi: 10.3390/cancers14143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheng G, Gao Y, Yang Y, Wu H. Osteosarcoma and Metastasis. Front Oncol. 2021;11:780264. doi: 10.3389/fonc.2021.780264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamplot J, Denduluri S, Qin J, Li R, Liu X, Zhang H, et al. The current and future therapies for human osteosarcoma. Curr cancer Therapy Reviews. 2013;9(1):55–77. doi: 10.2174/1573394711309010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris MA, Hawkins CJ. Recent and Ongoing Research into metastatic osteosarcoma treatments. Int J Mol Sci. 2022;23(7):3817–42. doi: 10.3390/ijms23073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neophytou CM, Panagi M, Stylianopoulos T, Papageorgis P. The role of Tumor microenvironment in cancer Metastasis: molecular mechanisms and therapeutic opportunities. Cancers. 2021;13(9):2053. doi: 10.3390/cancers13092053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidler IJ. The pathogenesis of cancer Metastasis: the’seed and soil’hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 10.Joyce JA, Pollard JW. Microenvironmental regulation of Metastasis. Nat Rev Cancer. 2009;9(4):239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sceneay J, Smyth MJ, Möller A. The pre-metastatic niche: finding common ground. Cancer Metastasis Rev. 2013;32(3):449–64. doi: 10.1007/s10555-013-9420-1. [DOI] [PubMed] [Google Scholar]
- 12.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17(5):302–17. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell. 2016;30(5):668–81. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Pan J, Barsky L, Jacob JC, Zheng Y, Gao C, et al. Characteristics of pre-metastatic niche: the landscape of molecular and cellular pathways. Mol Biomed. 2021;2(1):3. doi: 10.1186/s43556-020-00022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramani V, Teshima T, Tamura K, Chung J-S, Kobayashi M, Cruz PD, et al. Melanoma-Derived Soluble DC-HIL/GPNMB promotes Metastasis by excluding T-Lymphocytes from the pre-metastatic niches. J Invest Dermatology. 2018;138(11):2443–51. doi: 10.1016/j.jid.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Qi M, Xia Y, Wu Y, Zhang Z, Wang X, Lu L, et al. Lin28B-high Breast cancer cells promote immune suppression in the lung pre-metastatic niche via exosomes and support cancer progression. Nat Commun. 2022;13(1):897. doi: 10.1038/s41467-022-28438-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Z, Li Y-n, Jia S, Zhu M, Cao L, Tao M, et al. Lung mesenchymal stromal cells influenced by Th2 cytokines mobilize neutrophils and facilitate Metastasis by producing complement C3. Nat Commun. 2021;12(1):6202. doi: 10.1038/s41467-021-26460-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Han M, Gao J. Prognosis and targeting of pre-metastatic niche. J Controlled Release. 2020;325:223–34. doi: 10.1016/j.jconrel.2020.06.037. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Zhou L, Zhou J, Li Q, Ji Q. Underlying mechanisms and drug intervention strategies for the tumour microenvironment. J Experimental Clin Cancer Res. 2021;40(1):97. doi: 10.1186/s13046-021-01893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan TM, Roberts RD, Lizardo MM. Understanding and Modeling Metastasis Biology to improve therapeutic strategies for combating Osteosarcoma Progression. Front Oncol. 2020;10:13. doi: 10.3389/fonc.2020.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stastna M, Van Eyk JE. Secreted proteins as a fundamental source for biomarker discovery. Proteomics. 2012;12(4–5):722–35. doi: 10.1002/pmic.201100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue H, Lu B, Lai M. The cancer secretome: a reservoir of biomarkers. J Translational Med. 2008;6(1):52. doi: 10.1186/1479-5876-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns J, Wilding CP, Jones L, Huang RH. Proteomic research in sarcomas – current status and future opportunities. Sem Cancer Biol. 2020;61:56–70. doi: 10.1016/j.semcancer.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirikaew N, Pruksakorn D, Chaiyawat P, Chutipongtanate S. Mass Spectrometric-based proteomics for Biomarker Discovery in Osteosarcoma: current status and future direction. Int J Mol Sci. 2022;23(17):9741. doi: 10.3390/ijms23179741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerez S, Araya H, Thaler R, Charlesworth MC, López-Solís R, Kalergis AM, et al. Proteomic Analysis of Exosomes and Exosome-Free Conditioned Media from Human Osteosarcoma Cell Lines reveals secretion of proteins related to Tumor Progression. J Cell Biochem. 2017;118(2):351–60. doi: 10.1002/jcb.25642. [DOI] [PubMed] [Google Scholar]
- 26.Mazumdar A, Urdinez J, Boro A, Arlt MJ, Egli FE, Niederöst B, et al. Exploring the role of osteosarcoma-derived extracellular vesicles in pre-metastatic niche formation and Metastasis in the 143-B xenograft mouse osteosarcoma model. Cancers. 2020;12(11):3457. doi: 10.3390/cancers12113457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almeida SFF, Fonseca A, Sereno J, Ferreira HRS, Lapo-Pais M, Martins-Marques T et al. Osteosarcoma-Derived exosomes as potential PET imaging nanocarriers for lung Metastasis. Small. 2022:e2203999. [DOI] [PubMed]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–2. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 29.Morgan M, Anders S, Lawrence M, Aboyoun P, Pages H, Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25(19):2607–8. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC Bioinformatics. 2011;12(1):1–17. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickham H. ggplot2: Elegant graphics for Data Analysis.2016.
- 35.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. The Innovation. 2021;2(3):100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 37.Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc. 2019;14(1):68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 38.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and Metastasis. Nat Commun. 2020;11(1):1–19. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and Disease. Nat Rev Mol Cell Biol. 2014;15(12):786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popova NV, Jücker M. The functional role of extracellular matrix proteins in cancer. Cancers. 2022;14(1):238. doi: 10.3390/cancers14010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iqbal S, Leonard C, Regmi C, De Rantere S, Tailor D, Ren P. Lubricin/proteoglycan 4 binds to and regulates the activity of toll-like receptors in vitro. Sci Rep. 2016;6(1):1–12. doi: 10.1038/srep18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alquraini A, Garguilo S, D’Souza G, Zhang LX, Schmidt TA, Jay GD, et al. The interaction of lubricin/proteoglycan 4 (PRG4) with toll-like receptors 2 and 4: an anti-inflammatory role of PRG4 in synovial fluid. Arthritis Res Therapy. 2015;17(1):1–12. doi: 10.1186/s13075-015-0877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carpino G, Overi D, Melandro F, Grimaldi A, Cardinale V, Di Matteo S, et al. Matrisome analysis of intrahepatic cholangiocarcinoma unveils a peculiar cancer-associated extracellular matrix structure. Clin Proteomics. 2019;16(1):1–12. doi: 10.1186/s12014-019-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle ST, Poltavets V, Kular J, Pyne NT, Sandow JJ, Lewis AC, et al. ROCK-mediated selective activation of PERK signalling causes fibroblast reprogramming and tumour progression through a CRELD2-dependent mechanism. Nat Cell Biol. 2020;22(7):882–95. doi: 10.1038/s41556-020-0523-y. [DOI] [PubMed] [Google Scholar]
- 45.Bulla R, Tripodo C, Rami D, Ling GS, Agostinis C, Guarnotta C, et al. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun. 2016;7(1):1–11. doi: 10.1038/ncomms10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiffoleau E. C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front Immunol. 2018;9:227. doi: 10.3389/fimmu.2018.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, Xu C, Liu Z. S100 protein family in human cancer. Am J cancer Res. 2014;4(2):89. [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, Liu CH, Roberts AI, Das J, Xu G, Ren G, et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don’t know. Cell Res. 2006;16(2):126–33. doi: 10.1038/sj.cr.7310017. [DOI] [PubMed] [Google Scholar]
- 49.Marcuzzi E, Angioni R, Molon B, Calì B. Chemokines and chemokine receptors: orchestrating Tumor metastasization. Int J Mol Sci. 2018;20(1):96. doi: 10.3390/ijms20010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiratsuka S, Watanabe A, Sakurai Y, Akashi-Takamura S, Ishibashi S, Miyake K, et al. The S100A8–serum amyloid A3–TLR4 paracrine cascade establishes a pre-metastatic phase. Nat Cell Biol. 2008;10(11):1349–55. doi: 10.1038/ncb1794. [DOI] [PubMed] [Google Scholar]
- 52.Lee N-H, Park S-R, Lee JW, Lim S, Lee S-H, Nam S, et al. SERPINB2 is a novel indicator of cancer stem cell tumorigenicity in multiple cancer types. Cancers. 2019;11(4):499. doi: 10.3390/cancers11040499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Gui L, Zhang Y, Zhang J, Shi J, Xu G. Cystatin B is a progression marker of human epithelial ovarian tumors mediated by the TGF-β signaling pathway. Int J Oncol. 2014;44(4):1099–106. doi: 10.3892/ijo.2014.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aratani Y, Myeloperoxidase Its role for host defense, inflammation, and neutrophil function. Arch Biochem Biophys. 2018;640:47–52. doi: 10.1016/j.abb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S, Xia C, Xu C, Liu J, Zhu H, Yang Y, et al. Early growth response 3 inhibits growth of hepatocellular carcinoma cells via upregulation of Fas ligand. Int J Oncol. 2017;50(3):805–14. doi: 10.3892/ijo.2017.3855. [DOI] [PubMed] [Google Scholar]
- 55.Kaplan RN, Psaila B, Lyden D. Bone marrow cells in the ‘pre-metastatic niche’: within bone and beyond. Cancer Metastasis Rev. 2006;25(4):521–9. doi: 10.1007/s10555-006-9036-9. [DOI] [PubMed] [Google Scholar]
- 56.Yan J, Ye G, Shao Y. High expression of the ferroptosis-associated MGST1 gene in relation to poor outcome and maladjusted immune cell infiltration in uterine corpus endometrial carcinoma. J Clin Lab Anal. 2022;36(4):e24317. doi: 10.1002/jcla.24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu C, Qiang J, Deng Q, Xia J, Deng L, Zhou L, et al. ALDH1A1 activity in tumor-initiating cells remodels myeloid-derived suppressor cells to promote Breast cancer progression. Cancer Res. 2021;81(23):5919–34. doi: 10.1158/0008-5472.CAN-21-1337. [DOI] [PubMed] [Google Scholar]
- 58.Sokol CL, Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 2015;7(5):a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Qiang J, Yang X, Wang D, Rehman AU, He X, et al. IL1R2 blockade suppresses breast tumorigenesis and progression by impairing USP15-dependent BMI1 stability. Adv Sci. 2019;7(1):1901728. doi: 10.1002/advs.201901728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masucci MT, Minopoli M, Carriero MV. Tumor associated neutrophils. Their role in tumorigenesis, Metastasis, prognosis and therapy. Front Oncol. 2019;9:1146. doi: 10.3389/fonc.2019.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyagi A, Sharma S, Wu K, Wu SY, Xing F, Liu Y, et al. Nicotine promotes Breast cancer Metastasis by stimulating N2 neutrophils and generating pre-metastatic niche in lung. Nat Commun. 2021;12(1):474. doi: 10.1038/s41467-020-20733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ören B, Urosevic J, Mertens C, Mora J, Guiu M, Gomis RR, et al. Tumour stroma-derived lipocalin‐2 promotes Breast cancer Metastasis. J Pathol. 2016;239(3):274–85. doi: 10.1002/path.4724. [DOI] [PubMed] [Google Scholar]
- 63.Ren L, Mendoza A, Zhu J, Briggs JW, Halsey C, Hong ES, et al. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget. 2015;6(30):29469–81. doi: 10.18632/oncotarget.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jablonska J, Lang S, Sionov RV, Granot Z. The regulation of pre-metastatic niche formation by neutrophils. Oncotarget. 2017;8(67):112132–44. doi: 10.18632/oncotarget.22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan Z, Li Y, Zhang S, Wang X, Dou H, Yu X, et al. Extracellular matrix remodeling in Tumor progression and immune Escape: from mechanisms to treatments. Mol Cancer. 2023;22(1):48. doi: 10.1186/s12943-023-01744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mirzapour MH, Heidari-Foroozan M, Razi S, Rezaei N. The pro-tumorigenic responses in metastatic niches: an immunological perspective. Clin Transl Oncol. 2023;25(2):333–44. doi: 10.1007/s12094-022-02950-4. [DOI] [PubMed] [Google Scholar]
- 67.Li R, Wen A, Lin J. Pro-inflammatory cytokines in the formation of the pre-metastatic niche. Cancers. 2020;12(12):3752. doi: 10.3390/cancers12123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu J, Duan B, Jiang W, Fu S, Gao H, Lu L. Epidermal growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1) suppressed the growth of hepatocellular carcinoma cells by promoting semaphorin 3B(SEMA3B) Cancer Med. 2019;8(6):3152–66. doi: 10.1002/cam4.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic Metastasis. Nature. 2015;527(7578):329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeLeon-Pennell KY, Barker TH, Lindsey ML. Fibroblasts: the arbiters of extracellular matrix remodeling. Matrix Biol. 2020;91–92:1–7. doi: 10.1016/j.matbio.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao X, Chen J, Sun H, Zhang Y, Zou D. New insights into fibrosis from the ECM degradation perspective: the macrophage-MMP-ECM interaction. Cell & Bioscience. 2022;12(1):117. doi: 10.1186/s13578-022-00856-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeltz C, Primac I, Erusappan P, Alam J, Noel A, Gullberg D. Cancer-associated fibroblasts in desmoplastic tumors: emerging role of integrins. Sem Cancer Biol. 2020;62:166–81. doi: 10.1016/j.semcancer.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 73.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–98. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 74.Glabman RA, Choyke PL, Sato N. Cancer-Associated fibroblasts: Tumorigenicity and Targeting for Cancer Therapy. Cancers (Basel). 2022;14(16). [DOI] [PMC free article] [PubMed]
- 75.Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the Tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20(1):131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shani O, Raz Y, Monteran L, Scharff Ye, Levi-Galibov O, Megides O, et al. Evolution of fibroblasts in the lung metastatic microenvironment is driven by stage-specific transcriptional plasticity. eLife. 2021;10:e60745. doi: 10.7554/eLife.60745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pein M, Insua-Rodríguez J, Hongu T, Riedel A, Meier J, Wiedmann L, et al. Metastasis-initiating cells induce and exploit a fibroblast niche to fuel malignant colonization of the lungs. Nat Commun. 2020;11(1):1494. doi: 10.1038/s41467-020-15188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi X, Young CD, Zhou H, Wang X. Transforming growth Factor-β signaling in Fibrotic Diseases and Cancer-Associated fibroblasts. Biomolecules. 2020;10(12). [DOI] [PMC free article] [PubMed]
- 79.Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I et al. Autocrine TGF-β and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proceedings of the National Academy of Sciences. 2010;107(46):20009-14. [DOI] [PMC free article] [PubMed]
- 80.Mazumdar A, Urdinez J, Boro A, Migliavacca J, Arlt MJ, Muff R, et al. Osteosarcoma-Derived Extracellular vesicles induce lung fibroblast reprogramming. Int J Mol Sci. 2020;21(15):5451. doi: 10.3390/ijms21155451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung Metastasis. Nat Cell Biol. 2006;8(12):1369–75. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 82.Faria SS, Fernandes PC, Jr, Silva MJ, Lima VC, Fontes W, Freitas-Junior R, et al. The neutrophil-to-lymphocyte ratio: a narrative review. Ecancermedicalscience. 2016;10:702. doi: 10.3332/ecancer.2016.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma J, Kuzman J, Ray A, Lawson BO, Khong B, Xuan S, et al. Neutrophil-to-lymphocyte ratio (NLR) as a predictor for recurrence in patients with stage III Melanoma. Sci Rep. 2018;8(1):4044. doi: 10.1038/s41598-018-22425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu B, Huang Y, Sun Y, Zhang J, Yao Y, Shen Z, et al. Prognostic value of inflammation-based scores in patients with osteosarcoma. Sci Rep. 2016;6(1):39862. doi: 10.1038/srep39862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Granot Z, Henke E, Comen Elizabeth A, King Tari A, Norton L, Benezra R. Tumor Entrained neutrophils inhibit seeding in the Premetastatic Lung. Cancer Cell. 2011;20(3):300–14. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kowanetz M, Wu X, Lee J, Tan M, Hagenbeek T, Qu X, et al. Granulocyte-colony stimulating factor promotes lung Metastasis through mobilization of Ly6G + Ly6C + granulocytes. PNAS. 2010;107(50):21248–55. doi: 10.1073/pnas.1015855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Charan M, Dravid P, Cam M, Setty B, Roberts RD, Houghton PJ, et al. Tumor secreted ANGPTL2 facilitates recruitment of neutrophils to the lung to promote lung pre-metastatic niche formation and targeting ANGPTL2 signaling affects metastatic Disease. Oncotarget. 2020;11(5):510–22. doi: 10.18632/oncotarget.27433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Timpl R, Sasaki T, Kostka G, Chu ML. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol. 2003;4(6):479–89. doi: 10.1038/nrm1130. [DOI] [PubMed] [Google Scholar]
- 89.Zhang D, Han S, Pan X, Li H, Zhao H, Gao X, et al. EFEMP1 binds to STEAP1 to promote osteosarcoma proliferation and invasion via the Wnt/β-catenin and TGF-β/Smad2/3 signal pathways. J Bone Oncol. 2022;37:100458. doi: 10.1016/j.jbo.2022.100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Al Khader A, Fararjeh AFS, Kaddumi EG, Al-Saghbini M. Significance of fibulin-3 expression in Bladder cancer: a tissue microarray-based immunohistochemical study. World J Surg Oncol. 2022;20(1):1–6. doi: 10.1186/s12957-022-02597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen T-J, Chan T-C, Li W-S, Li C-F, Ke H-L, Wei Y-C, et al. Utility of EFEMP1 in the prediction of oncologic outcomes of urothelial carcinoma. Genes. 2021;12(6):872. doi: 10.3390/genes12060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu B, Thirtamara-Rajamani KK, Sim H, Viapiano MS. Fibulin-3 is uniquely upregulated in malignant gliomas and promotes Tumor cell motility and invasion. Mol Cancer Res. 2009;7(11):1756–70. doi: 10.1158/1541-7786.MCR-09-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Z, Cao C-J, Huang L-L, Ke Z-F, Luo C-J, Lin Z-W, et al. EFEMP1 promotes the migration and invasion of osteosarcoma via MMP-2 with induction by AEG-1 via NF-κB signaling pathway. Oncotarget. 2015;6(16):14191–208. doi: 10.18632/oncotarget.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in this study are available in the following databases:
RNA-Seq data: GEO database [GSE216744] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216744).
Protein identification: PRIDE database [PXD040814].