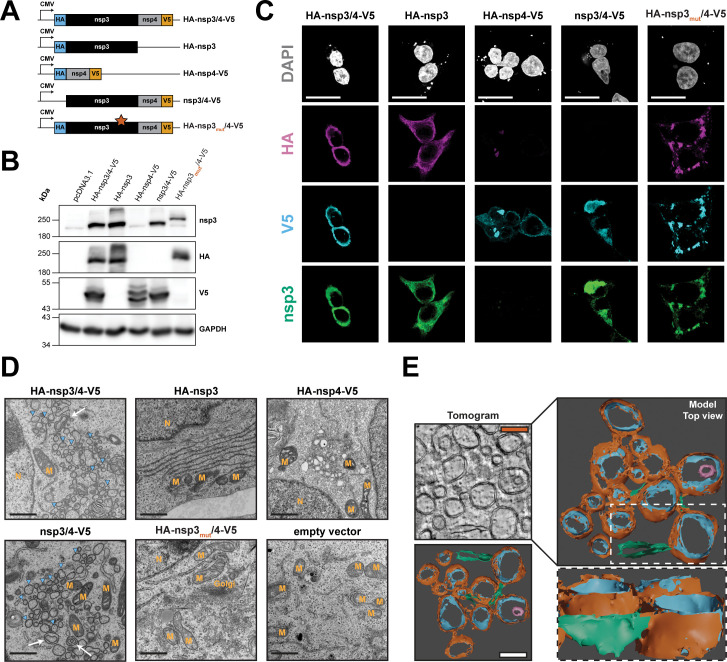
Fig 1.
Characterization of SARS-CoV-2 nsp3 and nsp4 in context of single and polyprotein expression. (A) Schematic overview of the expression constructs used in this study. Expression of the viral proteins is driven by the cytomegalovirus (CMV) immediate early promoter. The HA-nsp3mut/4-V5 construct contains the alanine substitution C1592A (star) that abrogates PLpro protease activity. (B) HEK293T cells were transfected with the given expression constructs, and 24 hours later, cells were lysed, and HA-nsp3- and nsp4-V5-tagged proteins were detected by western blot. GAPDH served as loading control; protein molecular mass in kilodalton is given on the left. (C) HEK293T cells grown on glass cover slips were transfected with the indicated constructs and fixed 24 hours post transfection. Immunostaining was performed with specific antibodies raised against nsp3, the HA epitope or the V5 epitope. Subcellular localization was visualized by confocal microscopy using a Leica SP8 confocal microscope and lightning deconvolution. Scale bars = 20 µm. (D) HEK293T cells were transfected with the indicated constructs and fixed with electron microscopy fixative 24 hours post-transfection before being processed for transmission electron microscopy analysis using a Jeol JEM-1400 electron microscope. Images of representative fields of view are shown. Blue arrowheads indicate DMVs, and white arrows indicate multimembrane vesicles. M, mitochondria; N, nucleus; Golgi, Golgi apparatus. Scale bars = 1 µm. (E) HEK293T cells were transfected with the HA-nsp3/4-V5 construct. After 24 hours, samples were fixed, embedded in epoxy resin, and a tilt-series was acquired using a FEI Tecnai F20. After alignment, 3D surface models were generated by segmentation in Ilastik and rendered using blender software. Scale bars = 250 nm. Orange, DMV outer membrane; blue, DMV inner membrane; magenta, inner multimembrane vesicle; green, ER.