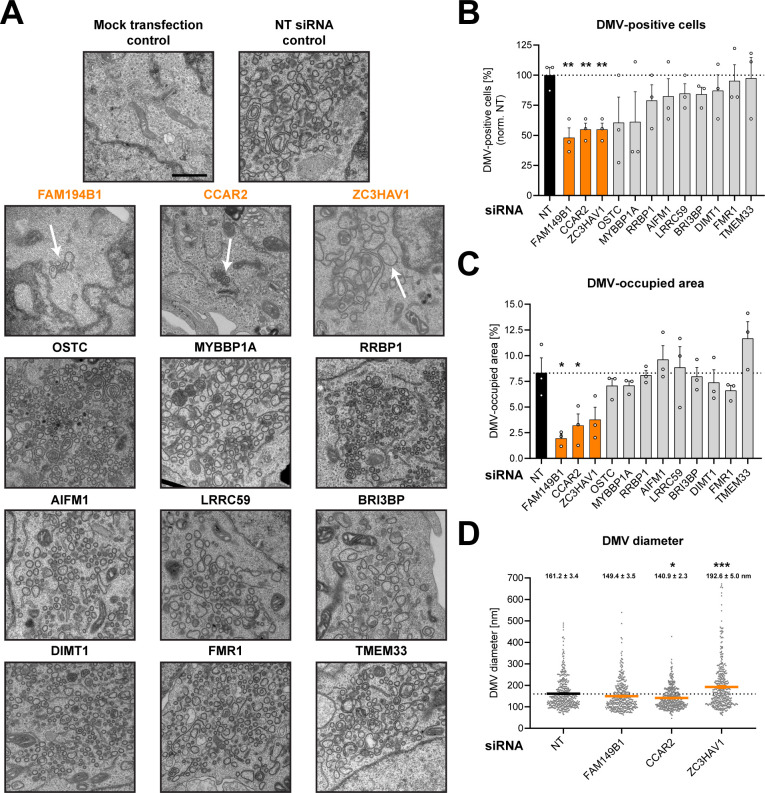
Fig 6.
FAM149B1, CCAR2, and ZC3HAV1 are host dependency factors involved in the biogenesis of SARS-CoV-2 nsp3/4-induced replication organelles. (A–D) HEK293T cells seeded onto coverslips in a 24-well plate were transfected with siRNA targeting the respective host dependency factors. And 48 hours post-transfection, the medium was replaced, and cells were transfected with the HA-nsp3/4-V5 expression construct. Also, 24 hours later, cells were fixed using EM fixative and processed for TEM. Overview images of the grid hexagons were taken at 200× magnification using SerialEM. Two micrographs per cell were acquired from all cells within the hexagon at a 10.000× magnification to assess DMV formation. (A) Representative images of DMVs are shown with white arrows indicating the few DMVs present in cells treated with siRNA targeting FAM149B1, CCAR2, and ZC3HAV1. DMV-positive cells (B) and the area occupied by DMVs (C) were quantified for at least 20 cells per replicate, and values were normalized to the NT control. (D) The diameter of DMVs present in cells treated with the NT siRNA control or with siRNAs targeting FAM149B1, CCAR2, and ZC3HAV1 was quantified for at least 75 DMVs per replicate. The experiments were performed in biological triplicates, and data are plotted as mean ± SEM. An unpaired t-test was performed to assess statistical differences with *P < 0.05, **P < 0.01, ***P < 0.001.