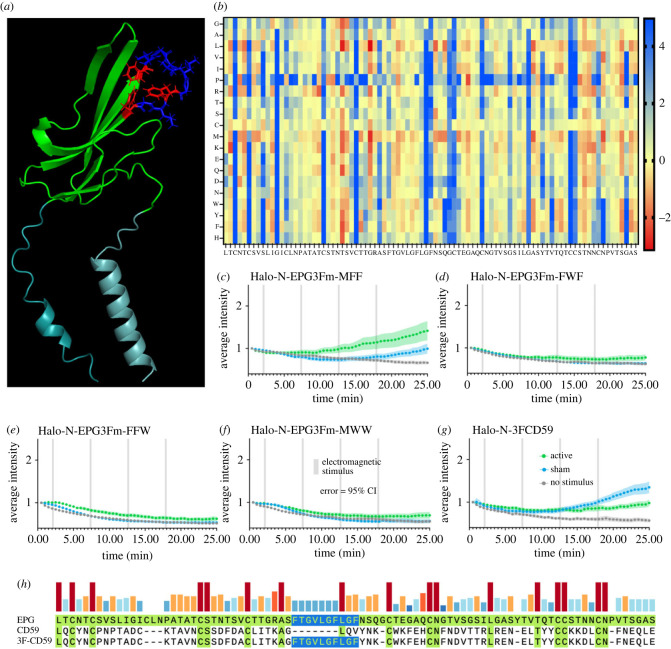
Figure 5.
Site directed mutagenesis of the three-phenylalanine region results in change of function. (a) Predicted structure of EPG with the three phenylalanine residues highlighted in red. (b) Stability of amino acid swaps for EPG in a heatmap; red is a stabilizing swap, yellow is neutral, and blue is a destabilizing swap. (c–g) Average intensity of GCaMP6m in HeLa cells expressing various 3F mutants over time with various stimuli. Error bars are representative of 95% CI. (c–f) Amino acids were swapped with the most stabilizing residue indicated in (b). (c) First F in 3F region knocked out. (d) Second F in 3F region knocked out. (e) Third F in 3F region knocked out. (f) All three F residues in 3F region knocked out. (g) 3F motif inserted into CD59. The Halo-N-EPG3Fm-MFF group included n = 77, n = 90, and n = 89 cells over 3 experiments for no stimulus, sham, and active groups respectively. The Halo-N-EPG3Fm-FWF group included n = 87, n = 96, and n = 86 cells over 3 experiments for the no stimulus, sham, and active groups respectively. The Halo-N-EPG3Fm-FFW group included n = 87, n = 85, and n = 88 cells over 3 experiments for the no stimulus, sham, and active groups respectively. The Halo-N-EPG3Fm-MWW group included n = 84, n = 83, and n = 89 cells over 3 experiments for the no stimulus, sham, and active groups respectively. The Halo-N-3FCD59 group included n = 86, n = 92, and n = 95 cells over 3 experiments for no stimulus, sham, and active groups respectively.