Fig. 5.
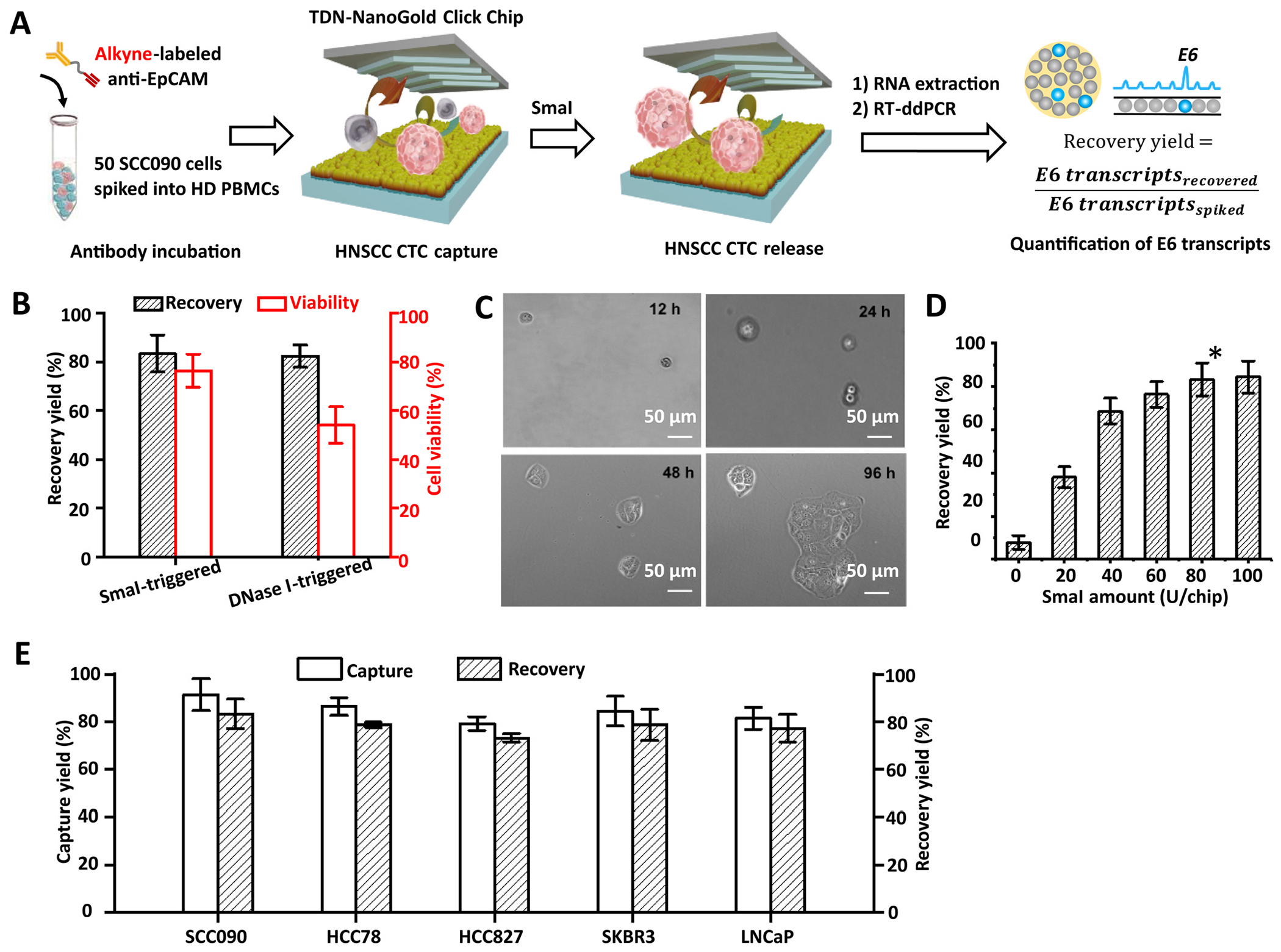
Optimization of TDN-NanoGold Click Chips for enzyme-triggered release of HNSCC CTCs. A) A general workflow for optimization of TDN-NanoGold Click Chips for enzyme-triggered release of HNSCC CTCs using artificial samples by RNA quantification. After CuAAC-mediated capture of HNSCC CTCs followed by their enzyme-triggered releases, E6 transcripts were quantified by RT-ddPCR, allowing calculation of recovery yields. B) Comparison of recovery yields and viabilities of CTCs recovered from the TDN-NanoGold Click Chips by employing specific restriction enzyme cleavage (SmaI; 80 U/chip) and non-specific endonuclease cleavage (DNase I; 60 U/chip). C) Micrographic images of the SCC090 cells for 12–96 h after their capture and release from TDN-NanoGold Click Chips. D) The recovery yields of TDN-NanoGold Click Chips according to the varying amounts of SmaI (0–100 U/chip) at a flow rate of 0.5 mL/h. * Indicates the optimal condition identified in this study. E) General applicability of the TDN-NanoGold Click Chips for CTC purification was validated by using artificial samples spiked with different types of cancer cells, i.e., SCC090 HNSCC cells, HCC78, and HCC827 lung cancer cells, SKBR3 breast cancer cells, and LNCaP prostate cancer cells (n = 3).
