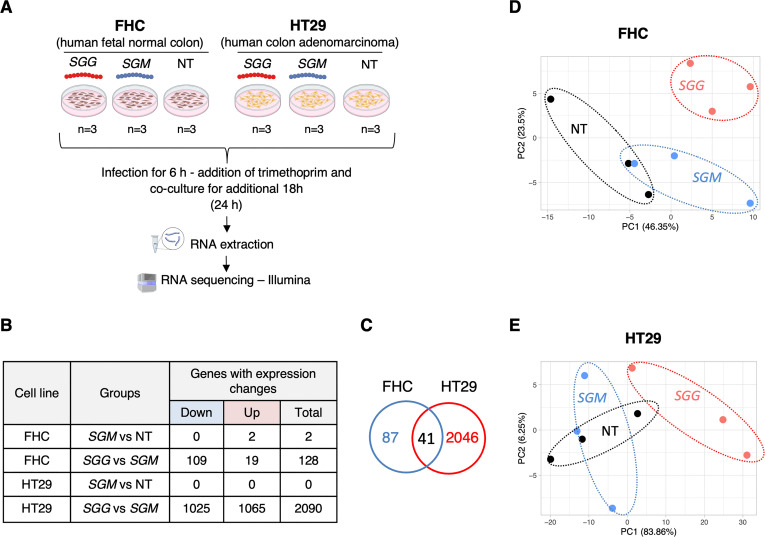
Fig 1. Comparison of host transcriptomic responses in normal FHC and tumoral HT29 colonic cells exposed to SGG vs SGM.
A. Experimental design of cell-bacteria co-culture used for transcriptomic study (created with BioRender.com). Cells were seeded onto 6-wells plate (FHC: 2 x 105 cells/well and HT29: 4 x 105 cells/well) and incubated for 16–20 hours. Cells were infected with bacteria: SGG UCN34 (6.5 x 104 CFU/ml) and SGM (6.5 x 105 CFU/ml). Trimethoprim (50 μg/ml) was added to the wells after 6 hours of co-culture to prevent bacterial over-growth. The total infection time was 24 h. Total RNA was prepared, processed and sequenced using Illumina technology. This experiment was performed in triplicate (n = 3). B. Table recapitulating the transcriptional changes in FHC and HT29 induced by SGM vs NT and SGG UCN34 vs SGM conditions. Criteria for significant gene changes were as follows: adjusted p < 0.05 and log2 fold change (FC) > 0.5 or < -0.5). C. Venn diagram showing that 41 genes regulated upon SGG UCN34 infection were in common between FHC and HT29 cells. D. PCA of transcriptome samples in FHC cells: SGG (n = 3); SGM (n = 3) and NT (n = 3). E. PCA of transcriptome samples in HT29 cells: SGG (n = 3); SGM (n = 3) and NT (n = 3). D and E. Each PCA is based on the variance-stabilized transformed count matrix that has been adjusted for the replicate effect using the remove BatchEffect function or the limma R package (version 3.52.4).