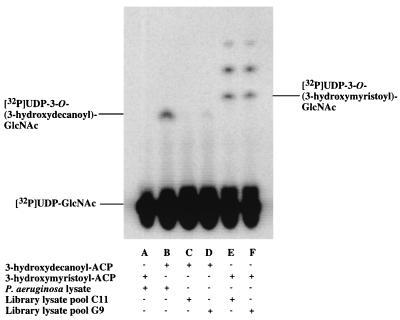
FIG. 2.
Expression cloning of a 3-hydroxydecanoyl-specific UDP-GlcNAc acyltransferase from a P. aeruginosa PAO1 DNA library in E. coli LE392 assayed in pools of four. When [α-32P]UDP-GlcNAc is acylated, it migrates off the origin in this thin-layer system, as indicated. Acylation with 3-hydroxymyristate results in a product that migrates slightly faster than does the product of acylation with 3-hydroxydecanoate. Extracts of wild-type P. aeruginosa acylate [α-32P]UDP-GlcNAc efficiently in the presence of 3-hydroxydecanoyl–ACP (lane B) but not at all in the presence of 3-hydroxymyristoyl–ACP (lane A). Conversely, strains of E. coli that do not harbor the P. aeruginosa lpxA gene acylate [α-32P]UDP-GlcNAc in the presence of 3-hydroxymyristoyl–ACP (lane E) but not in the presence of 3-hydroxydecanoyl–ACP (lane C). The more rapidly migrating compounds in lanes E and F represent further metabolites of the lipid A pathway that are derived in E. coli extracts from acylated-UDP-GlcNAc (1, 3). Lanes C to F show the assay results obtained with pools of four lysates (prepared as described in Materials and Methods), each derived from distinct E. coli colonies harboring different P. aeruginosa DNA inserts. With 3-hydroxydecanoyl–ACP as the substrate, only lysate pool G9 (of the 288 pools assayed) catalyzed measurable acylation of [α-32P]UDP-GlcNAc (lane D). With 3-hydroxymyristoyl–ACP as the substrate (lane F), pool G9 was comparable in activity to all other pools in the collection (such as C11 in lane E), indicating that the protein concentrations of G9 and C11 were about the same.