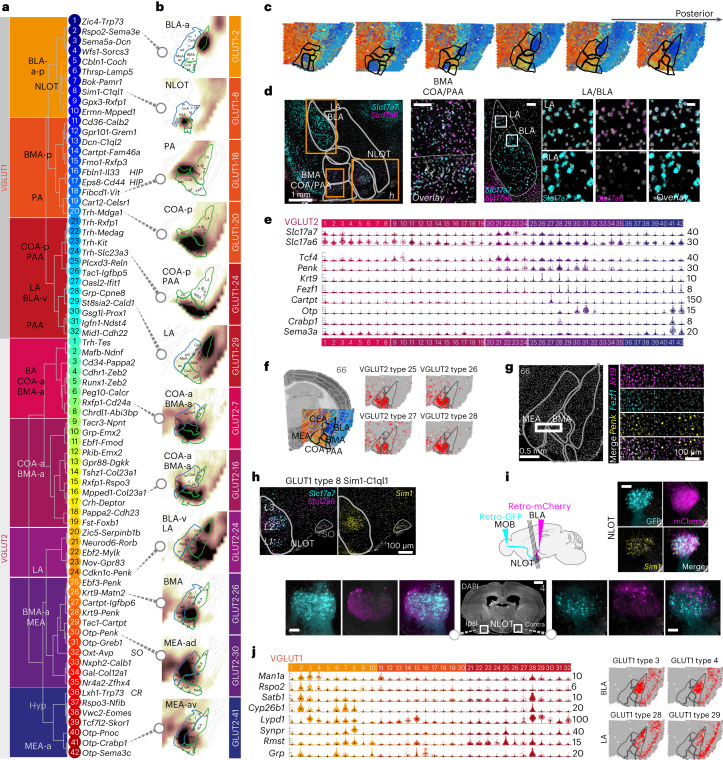
Fig. 3. Glutamatergic diversity.
a, Dendrogram of glutamatergic cell types of the VGLUT1 and VGLUT2 classes, with cluster number and two-gene identifier; likely location indicated on the left. b, Examples of inferred spatial distributions of 12 distinct cell types in a relevant coronal section with high correlation to AMBA. Correlation coefficients visualized as a heatmap (white, low and brown, high correlation). c, Weighted spatial correlation of GABA types to six coronal sections Visium ST. Each dot represents one capture spot, colored by the dominant cell type (see a, right). d, Multiplexed fluorescent in situ hybridization of Slc17a7 (VGLUT1) and Slc17a6 (VGLUT2) highlighted in LA, BLA (scale bar, 20 μm) and anterior COA/BMA (scale bar, 100 μm) in a single representative anterior section. Top panel section overview: scalebars, 500 μm; LA/BLA overview: scale bar, 100 μm. e, VGLUT2 MEA/BMA gene expression visualized as single-cell violin plots. Each dot represents one cell; black square represents median cluster expression and the number of molecules (y scale maximum) is on the right. f, Examples of related BMA/MEA populations visualized as correlation heatmap to Visium ST (gray, low and red, high). g, Multiplex fluorescence in situ hybridization for Krt9+ MEA/BMA populations, some Penk+ and coexpressing Fezf1 in the MEA only. h, Multiplex FISH validation of Slc17a7+Slc17a6+Sim1+ cells in the NLOT (overview section in c); Scale bar, 100 μm. i, NLOT-projection tracing using retro-AAVs to the MOB (hSyn1-EGFP) and BLA (hSyn1-mCherry) reveals NLOT cell labeling ipsi- and contralateral to the injection sites, of Sim1+ cells (multiplex FISH). Scale bars, overview, 1 mm; zoom-ins, 100 mm. j, Left, BLA/LA cell types gene expression; right, examples of spatial distributions. Gene expression visualized as single-cell violin plots, each dot represents one cell; black square represents median expression and number of molecules (y-scale maximum) are on the right. BLA/LA populations distribution visualized as correlation heatmap to Visium ST (gray, low and red, high). SO, supraoptic nucleus.