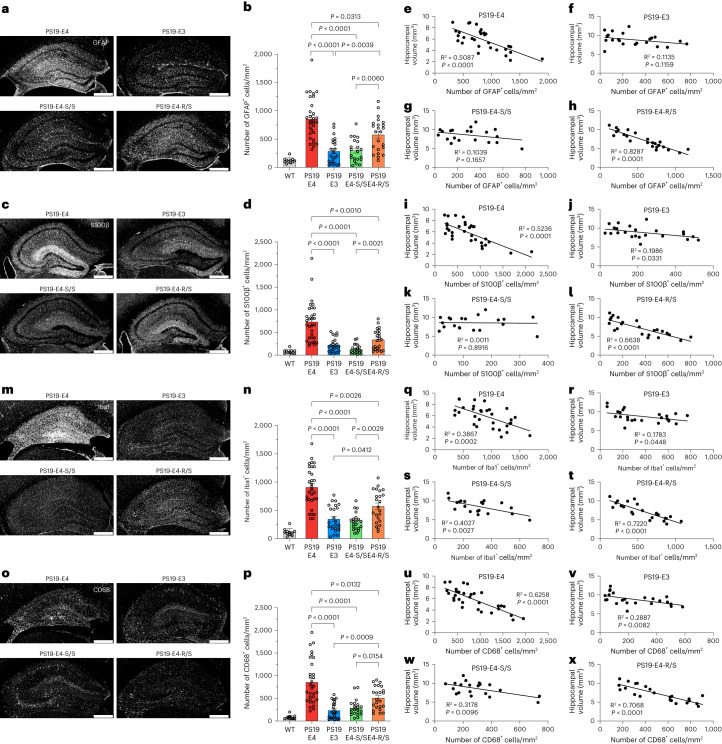
Fig. 5. The R136S mutation reduces APOE4-driven gliosis in tauopathy mice.
a,b, Representative images of GFAP immunostaining of astrocytes in the hippocampus of 10-month-old PS19-E4, PS19-E3, PS19-E4-S/S and PS19-E4-R/S mice (a) and quantification of the number of GFAP+ cells per mm2 (b) in the hippocampus of these mice and WT mice. c,d, Representative images of S100β immunostaining of astrocytes in the hippocampus of 10-month-old PS19-E4, PS19-E3, PS19-E4-S/S and PS19-E4-R/S mice (c) and quantification of the number of S100β+ cells per mm2 (d) in the hippocampus of these mice and WT mice. e–h, Correlations between GFAP+ cells per mm2 and hippocampal volume in PS19-E4 (n = 31) (e), PS19-E3 (n = 23) (f), PS19-E4-S/S (n = 20) (g) and PS19-E4-R/S (n = 23) (h) mice. i–l, Correlations between S100β+ cells per mm2 and hippocampal volume in PS19-E4 (n = 31) (i), PS19-E3 (n = 23) (j), PS19-E4-S/S (n = 20) (k) and PS19-E4-R/S (n = 23) (l) mice. m,n, Representative images of Iba1 immunostaining of microglia in the hippocampus of 10-month-old PS19-E4, PS19-E3, PS19-E4-S/S and PS19-E4-R/S mice (m) and quantification of the number of Iba1+ cells per mm2 (n) in the hippocampus of these mice and WT mice. o,p, Representative images of CD68 immunostaining of microglia in the hippocampus of 10-month-old PS19-E4, PS19-E3, PS19-E4-S/S and PS19-E4-R/S mice (o) and quantification of the number of CD68+ cells per mm2 (p) in the hippocampus of these mice and WT mice. q–t, Correlations between Iba1+ cells per mm2 and hippocampal volume in PS19-E4 (n = 31) (q), PS19-E3 (n = 23) (r), PS19-E4-S/S (n = 20) (s) and PS19-E4-R/S (n = 23) (t) mice. u–x, Correlations between CD68+ cells per mm2 and hippocampal volume in PS19-E4 (n = 31) (u), PS19-E3 (n = 23) (v), PS19-E4-S/S (n = 20) (w) and PS19-E4-R/S (n = 23) (x) mice. For all quantifications in b,d,n,p, WT, n = 11; PS19-E4, n = 31; PS19-E3, n = 24; PS19-E4-S/S, n = 21; PS19-E4-R/S, n = 23. Scale bars, 500 µm in a,c,m,o. Throughout, data are expressed as mean ± s.e.m. In b,d,n,p, differences between groups were determined by Welch’s ANOVA followed by Dunnett’s T3 multiple comparison test. Comparisons of P ≤ 0.05 are labeled on the graph. Pearson’s correlation analysis (two-sided).